Statistical Approach for Optimization of Independent Variables on Alkali-Thermo Stable Protease Production from Bacillus Licheniformis Strain BIHPUR 0104
Ameer Khusro
Department of Plant Biology and Biotechnology,Loyola College,Nungambakkam,Chennai,Tamil Nadu,India
Received date: October 15,2015; Accepted date: October 26,2015; Published date: October 29,2015
Abstract
An alkalophilic and thermostable protease producing Bacillus licheniformis strain BIHPUR 0104 was isolated from poultry farm. Response surface methodology by Box-Behenken design was employed for the enhanced production of protease using independent variables selected through one variable at one time approach. Under the optimized condition of sucrose (1.5% w/v), temperature (40°C) and incubation period (24 h), the protease production from strain BIHPUR 0104 was enhanced 2 fold compared to one variable at one time approach. The protease experimental yield (483.23 U/ml) closely matched the predicted value (480.74 U/ml) by the statistical model (R2=0.9997). The p values showed that the independent variables had highly significant effect on proteolytic activity (p<0.0001) of the strain tested. The crude protease obtained from strain BIHPUR 0104 showed stability at a wide range of pH (9) and temperature (50°C) indicating thermo-alkali stable nature of the crude enzyme.
Keywords
Bacillus licheniformis; Box-Behenken design; Protease; Poultry farm; Response surface methodology.
Introduction
Proteases are one of the most important groups of enzymes whose two-third of the production depends upon bacterial origins. Among proteases,alkaline proteases are extensively used in leather,detergent,food,pharmaceutical,textile,organic chemical synthesis,waste water treatment,and other industries [1]. Due to growing demand of alkaline protease at industrial scale,biotechnologists are looking for novel sources of proteases. The successful exploitation of microbial diversity for targeting new proteases with industrially desirable attributes has been a continuous effort [2]. Bacteria are ubiquitous in nature and their enzymes are potentially exploited for several industrial purposes leading to development of environment friendly process. Of these bacterial sources,commercial production of alkaline protease at large scale from Bacillus sp. dominates in industries. The optimization of fermentation conditions is the most important step in the development of a cost-effective fermentation process from bacterial sources. Several physiochemical parameters and biological factors influence the production of enzymes from bacteria. Hence,it is important to optimize the medium components and various physiochemical factors influencing the enzyme production,through systematic and sequential optimization strategy. ‘One-variable-at-a-time’ (OVAT) conventional approach of optimization is a time restrictive and expensive process that does not establish combined interaction(s) among various variables [3]. Statistical optimization tools such as Box-Behenken design (BBD) of response surface methodology (RSM),is widely used for process optimization by generating empirical models [4]. Considering the wide range production of proteases from Bacillus sp. and economic statistical optimization approach using RSM,the present investigation was carried out to isolate and optimize independent variables influencing alkaline and temperature protease production using BBD design from poultry farm associated Bacillus sp.
Methods
Chemicals,media and analysis
All the chemicals and media used in the present investigation wereof analytical grade obtained from Sigma-Aldrich Chemicals Ltd (St. Louis MO,USA) and HiMedia Laboratories Ltd (Mumbai,India). The statistical software package “Design Expert” 9.0.0 (Stat-Ease Inc.,Minneapolis,USA) was used to analyze the experimental design and the regression analysis of the experimental data.
Isolation and characterization of proteolytic bacterium
Poultry feces soil sample was serially diluted and 0.1 of the inoculum was streaked on skim milk agar plate (skim milk 1 g; agar 1.8 g; distilled water 100 ml). The colonies showing zone of hydrolysis on agar plate were selected and extracellular protease production by the isolates were observed on skim milk agar plate after centrifuging and collecting the supernatant of the overnight grown cultures. The bacterium showing maximum zone of hydrolysis on skim milk agar plate was selected and characterized further.
Isolate showing potential protease production was characterized by cultural,morphological and biochemical analysis using Bergey’s Manual of Systemic Bacteriology. Genomic DNA of the bacteria was isolated and the amplification of the 16S ribosomal RNA was done using the PCR (ependorfep.Gradient) with Taq DNA polymerase and primers 27F (5` AGTTTGATCCTGGCTCAG 3`) and 1492R (5`ACGGCTACCTTGTTACGACTT 3`). The conditions for thermal cycling were as follows: denaturation of the target DNA at 94°C for 4 min followed by 30 cycles at 94°C for 1 min,primer annealing at 52°C for 1 min and primer extension at 72°C for 1 min. At the end of the cycling,the reaction mixture was held at 72°C for 10 min and then cooled to 4°C. PCR amplification was detected by agarose gel electrophoresis and visualized by gel doc. The amplicon obtained was sequenced by an automated sequencer (Genetic Analyzer 3130,Applied Biosystems,and USA) and the sequences were compared for similarity search with the reference species of bacteria contained in genomic database banks,using the NCBI BLAST.
Protease Assay
Extracellular protease activity was measured using casein as a substrate [5]. An assay mixture containing 0.5 ml of substrate (1% casein in Tris– HCl buffer,50mM,pH 9.0) and 0.5 ml of crude enzyme was incubated at 50°C for 20 min. Reaction was stopped with 2.5 ml of 5% trichloroacetic acid,and the contents were centrifuged at 10,000 rpm for 20 min at 4°C. The absorbance was examined spectrophotometerically at 280 nm with the standard curve of tyrosine. One unit of enzyme activity is defined as the amount of enzyme required to release 1 mg tyrosine from casein per minute under assay conditions.
Optimization of process parameters for protease production using OVAT method
Effect of various process parameters on extracellular protease production by the isolate was estimated under submerged cultivation. The parameters optimized using OVAT method were pH (5-10),temperature (30°C - 50°C),agitation speed (120 – 160 rpm),inoculum size (0.25% - 2%),carbon sources (glucose,casein,xylose,sucrose,lactose and mannose) and nitrogen sources (peptone,yeast extract,beef extract,tryptone,KNO3,ammonium sulphate and ammonium chloride). The protease activity was estimated as described earlier.
Effect of pH and temperature on protease stability
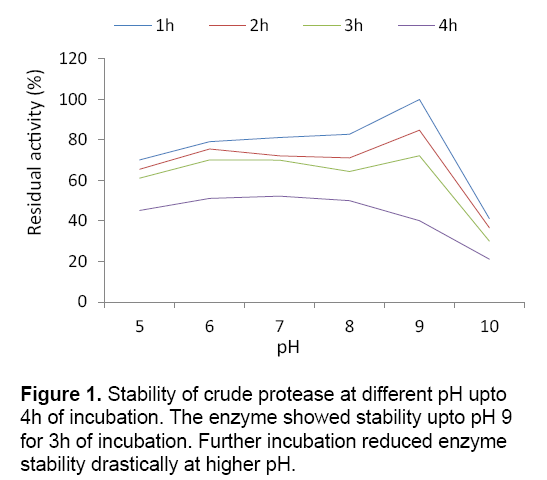
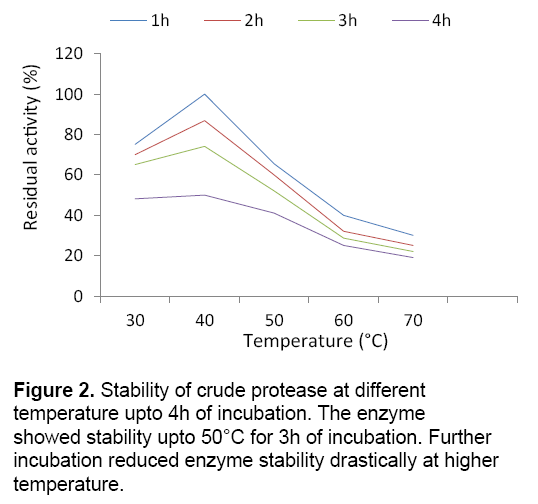
Stability of crude enzyme was tested by incubating in buffers of different pH (5–10) at 35°C upto 4h and residual activity was determined after each 1h of incubation under standard assay condition. The thermal stability was studied by incubating enzymes at various temperatures (30–70°C) and residual activity was measured after each 1h of interval up to 4h under standard assay condition.
Process optimization of independent variables using RSM
Based on the OVAT results,the independent variables such as sucrose,incubation temperature and incubation period were found to have profound effect on protease production. These independent factors were chosen and optimized by RSM using Box-Behnken design. Each variable was studied at three different levels (-1,0,+1). A mathematical correlation among the selected variables on protease production was developed and the data obtained from the design were fitted to a second-order polynominal equation. The protease activity (Y) was describes as a function of the independent variables.
Y = β0 + ? βiXi + ? βiiXi2 + ? βijXiXj
where,Y = predicted response
β0 = intercept term
βi= linear effect
βii= squared effect
βij= interaction effect
Xi,Xj = independent variables
The goodness of fit of the polynomial equation was expressed by coefficient of determination R2 and its statistical significance level was checked by F test.
Model validation
The statistical model was validated with respect to all the three independent variables within the design space by selecting randomly the experimental runs and estimating protease production for the same.
Results and Discussion
Isolation and screening of protease producing bacteria
Five bacteria were isolated from feces soil samples of poultry farm,collected from Guduvanchery. These isolates were further screened for extracellular protease production. Of 5 isolates,1 showed the production of protease as indicated by appearance of clear halo zone around the agar wells at 37°C (Figure not shown). This protease positive isolate was further selected for cultural,morphological and biochemical characteristics.
Identification and characterization of bacteria
The cultural,morphological and biochemical characteristics of the isolate was studied (Data not shown). All the bacterial cultures showed white and spreading type colonies. The microscopic observation showed the morphology of cultures as rod shaped with endospore. The isolated bacterial strain was identified as Bacillus sp. based on the taxonomical characteristic.
Genomic DNA of the isolate was visualized under UV. The amplicon of 712 bp sequences (KC424492) was observed using PCR amplification (Figure not shown). In the present study,16S rRNA gene sequencing of the isolate was investigated. The isolate was identified as Bacillus licheniformis strain BIHPUR 0104 (KC424492) by comparing the similarity with the reference species of bacteria contained in genomic database banks,using the NCBI BLAST. The comparison showed that the similarities of 16S rRNA gene sequences were 99%. The identity of strain was determined by comparing them with the available sequences of the strains and with high scored rRNA sequences in BLAST search.
Process optimization using OVAT method
Submerged fermentation (SmF) is primarily used for the production of industrially important enzymes from various bacterial sources. The bacterium showed maximum protease production (150.16 U/ml) at pH 9 under submerged cultivation. Lower and higher pH of the medium showed adverse effect on enzyme production. The present study favours the finding of Vijayaraghavan et al. [6] who estimated maximum protease production from Bacillus sp. at pH 9. The reduction in protease production at lower and higher pH may be due to the suppression of metabolic activity of bacterium at higher or lower pH,thus decreasing the enzyme production. In the present context,the crude protease obtained from the strain showed stability upto pH 9 for 3 h of incubation with residual activity of 40.12%. (Figure 1) The result determined the alkali tolerant nature of the protease obtained from BIHPUR 0104.
Temperature is one of the important criteria in order to enhance enzyme production in bacteria under submerged cultivation. Strain BIHPUR 0104 showed maximum protease activity (175.11 U/ml) at 35°C,representing the bacteria mesophilic in nature. According to the report of Singh and Bajaj [7],Bacillus sp. showed maximum protease activity at 60°C. In the present investigation,stability of the crude protease at higher temperature (50°C) is the unique characteristic of the isolate (Figure 2 ). The protease obtained from the isolate showed stability at high temperature with 41.32% of residual activity upto 3h of incubation,indicating thermostable characteristic of the enzyme.
Aeration is one of the important parameters affecting the rate of enzyme production in bacteria. In the present investigation,the rate of protease production from the strain BIHPUR 0104 was increased at 130 rpm with maximum activity of 185.14 U/ml. The agitation rate lower and higher than 130 rpm showed moderate effect on the protease activity. On the other hand the protease activity was found to be high (198.23 U/ml) after 24 h of incubation and the enzyme activity decreased considerably. There are reports on variation in maximum enzyme production time among different Bacillus sp.[8,9] and depends on the bacterial strain type,cultural and environmental conditions,and the genetic potential of the bacteria.
Carbon sources in simpler form are the source of growth and metabolism in bacteria. Of the different carbon sources tested for the profound effect on the protease production,sucrose achieved maximum protease productivity of 235.18 U/ml. The results totally agree with the findings of Vijayaraghavan et al [6]. Furthermore,the supplemented nitrogen sources had moderate effect on protease productivity. The results showed that there was a slight increase in alkaline protease production with various tested nitrogen sources. Among the nitrogen sources,yeast extract showed the maximum production of protease (239.15 U/ml) than other nitrogen sources. The investigation is in good agreement with the finding of Prakasham et al. [10] who demonstrated yeast extract as a potential nitrogen source for the maximum productivity of protease.
Optimization of variables through response surface methodology approach
Based on the OVAT results,independent variables such as sucrose (1% w/v),temperature and incubation period were found to be effective factors influencing the alkaline and temperature stable protease production. The independent variables were selected for further optimization through BBD using RSM approach. The selected variables with their respective high and low concentrations used in optimization process are represented in Table 1. To optimize carbon source,temperature and incubation period,a BBD design,consisting of a set of 17 experiments with three levels of three variables were conducted. The BBD matrix of the independent variables in coded units along with predicted and experimental values of response is given in Table 2.
| Variables | Code | Range and levels | ||
| -1 | 0 | +1 | ||
| Sucrose (% w/v) | A | 0.5 | 1 | 1.5 |
| Temperature (°C) | B | 30 | 35 | 40 |
| Incubation period (h) | C | 12 | 24 | 48 |
Table 1: Experimental range, level and code of independent variables for BBD design.
| Run Order | A | B | C | Protease activity (U/ml) | |
|---|---|---|---|---|---|
| Experimental | Predicted | ||||
| 1 | +1 | -1 | 0 | 313.63 | 314.09 |
| 2 | 0 | -1 | +1 | 182.35 | 180.86 |
| 3 | -1 | -1 | 0 | 369.12 | 371.61 |
| 4 | +1 | 0 | -1 | 224.43 | 225.43 |
| 5 | 0 | 0 | 0 | 203.36 | 201.96 |
| 6 | -1 | 0 | -1 | 204.45 | 203.43 |
| 7 | 0 | +1 | +1 | 269.19 | 270.66 |
| 8 | 0 | 0 | 0 | 202.13 | 201.96 |
| 9 | 0 | 0 | 0 | 200.87 | 201.96 |
| 10 | +1 | 0 | +1 | 280.78 | 281.80 |
| 11 | -1 | 0 | +1 | 204.28 | 203.28 |
| 12 | 0 | 0 | 0 | 203.23 | 201.96 |
| 13 | 0 | -1 | -1 | 185.14 | 183.67 |
| 14 | 0 | 0 | 0 | 200.23 | 201.96 |
| 15 | 0 | +1 | -1 | 210.14 | 211.63 |
| 16 | -1 | +1 | 0 | 323.16 | 322.70 |
| 17 | +1 | +1 | 0 | 483.23 | 480.74 |
Table 2: Box-Behenken design along with experimental and predicted values of response.
The production of protease (Y) was predicted by the following model:
Y (U/ml) = 201.96 + 25.13A + 29.44B + 14.06C + 53.89AB + 14.13AC + 15.46BC + 93.55A2 + 76.77B2 – 67.03C2
Highest protease activity was seen with run order number 17 using sucrose (1.5% w/v),temperature (40°C) and incubation period (24 h). The maximum experimental protease activity by the strain was estimated 483.23 U/ml with the predicted value of 480.74 U/ml.
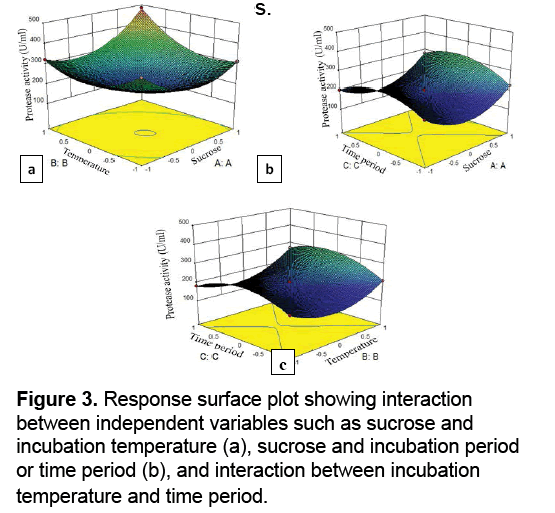
Response surface plots were constructed for the identification of interdependent correlation between the variables by constructing the protease activity on the z-axis against any two independent variables. Figure 3 a-c represents the 3D surface plots among the variables for the response. The shape of response graph represents the interaction between Analysis of variance (ANOVA) for protease quadric model is given in Table 3. The p value is used for checking the significance of each of the coefficient. The multiple correlation coefficients (R2) closer to 1 denote better correlation between experimental and predicted response values. The model can be used to navigate the design space. The experimental values of protease activity were in close agreement with the statistically predicted ones,confirming the validation of model for the optimization of the independent variables.
| Source | Sum of Squares |
df | Mean Square |
F- Value | p-value | |
|---|---|---|---|---|---|---|
| Prob> F | ||||||
| Model | 1.060E+005 | 9 | 11777.76 | 2469.15 | < 0.0001 | significant |
| A-A | 5053.14 | 1 | 5053.14 | 1059.37 | < 0.0001 | |
| B-B | 6931.35 | 1 | 6931.35 | 1453.12 | < 0.0001 | |
| C-C | 1580.34 | 1 | 1580.34 | 331.31 | < 0.0001 | |
| AB | 11616.53 | 1 | 11616.53 | 2435.35 | < 0.0001 | |
| AC | 798.63 | 1 | 798.63 | 167.43 | < 0.0001 | |
| BC | 956.05 | 1 | 956.05 | 200.43 | < 0.0001 | |
| A2 | 36849.25 | 1 | 36849.25 | 7725.26 | < 0.0001 | |
| B2 | 24815.62 | 1 | 24815.62 | 5202.47 | < 0.0001 | |
| C2 | 18917.70 | 1 | 18917.70 | 3966.00 | < 0.0001 | |
| Residual | 33.39 | 7 | 4.77 | |||
| Lack of Fit | 25.61 | 3 | 8.54 | 4.39 | 0.0936 | not significant |
| Pure Error | 7.78 | 4 | 1.95 | |||
| Cor Total | 1.060E+005 | 16 |
R2: 0.9997, adj R2: 0.9993, predicted R2: 0.9960
CV: 0.87%, adeq precision: 179.024
df = degree of freedom
Table 3: Analysis of variance (ANOVA) for protease activity.
Conclusion
The application of statistical based approach for process optimization of enzyme production has gained increased importance due to its easy applicability and validity. The present investigation established that Bacillus licheniformis strain BIHPUR 0104 isolated from poultry farm produces alkalithermo stable proteases efficiently under submerged cultivation. Optimization of independent variables such as sucrose (1.5% w/v),temperature (40°C) and incubation period (24 h) through BBD using RSM showed 2 fold enhancements in protease production compared to OVAT method. The strain BIHPUR 0104 can be successfully utilized at industrial scale for enhanced alkali-thermo stable protease production using statistical approach.
References
- Shah K,Mody K,Keshri J,Jha B. (2010). Purification and characterization of a solvent,detergent and oxidizing agent tolerant protease from Bacillus cereus isolated from the Gulf of Khambhat. J. Mol. Catal. B. 67: 85-91.
- Joo HS,Chang CS. (2005). Oxidant and SDS stablealkaline protease from a halotolerantBacillus clausii I-52: enhanced production and simple purification. J. Appl. Microbiol. 98: 491–497.
- Vishwanatha KS,Rao AGA,Singh SA. (2010). Acid protease production by solid state fermentation using Aspergillusoryzae MTCC 5341: optimization of process parameters. J. Ind. Microbiol. Biotechnol37: 129–138.
- Asif M,Hussain A,Ali MA,Rasool M. (2012). Scale upstudies for the production of protease enzyme using Bacillus subtilis adopting response surface methodology. African J. Microbiol Res. 6: 2120–2128.
- Kembhavi AA,Kulkarni A,Pant A. (1993). Salt-tolerant and thermostable alkaline protease from Bacillus subtilis NCIM No. 64. Appl. Biochem. Biotechnol38: 83–92.
- Vijayaraghavan P,Kalaiyarasi M,GnanaPrakash Vincent S. (2015).Statistical approach for the production and partial characterization of alkaline stable protease from a newly isolated Bacillus sp. IND6 for silver recovery. Res. J. Microbiol. 10: 83-99.
- Singh S,Bajaj BK. (2015). Medium optimization for enhanced production of protease with industrially desirable attributes from Bacillus subtilis K-1. Chem. Eng. Commun. 202: 1051-1060
- Hindhumathi M,Vijayalakshmi S,Thankamani V. (2011). Optimization and cultural characterization of alkalophilic protease producing Bacillus sp. GPA4. Res. Biotechnol. 2: 13–19.
- Bajaj BK,Manhas K. (2012). Production and characterization of xylanase from Bacillus licheniformis P11(C) with potential for fruit juice and bakery industry. Biocatal. Agric. Biotechnol. 1: 330–337.
- Prakasham RS,SubbaRao CH,Sarma PN. (2006). Green gram husk: An inexpensive substrate for alkaline protease production by Bacillus sp. in solid-state fermentation. Bioresource. Technol. 97: 1449-1454.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences