Binding of EGFR Gene Fragments to Nuclear Matrix Proteins in Bladder Cancer
Shudao Xiong, Bogui Wen, Jingrong Li, Guanwu Li, Jing Li
1Institute of Medical Science, the First Affiliated Hospital of Wenzhou Medical College, Wenzhou 325000, China
2Department of Molecular Biology, Shantou University Medical College, Shantou 515031, China
Abstract
This paper studied correlation between EGFR gene fragments and nuclear matrix proteins (NMPs) in human bladder transitional cell carcinoma (hBTCC). Both genomic DNA and nuclear matrix DNA (H0DNA, H25DNA, H50DNA, H100DNA) of human bladder carcinoma could amplify 110bp positive fragment with primers II even DNase I concentration was up to 100 IU/ml. Two pair primers were synthesized according to the cDNA sequence of EGFR gene and two EGFR fragments were amplified by PCR using the two pair primers in genomic DNA and different nuclear matrix DNA. Southwestern blot analysis demonstrated that EGFR gene fragment (3901-4010nt) was bound to a about 60kD nuclear matrix protein in both NMPs (tissues and T24 cells) and total cell protein of bladder carcinoma tissues, but not in cytoplasm sample.
Keywords
EGFR, nuclear matrix proteins, bladder transitional cell carcinoma, PCR, southwestern blot
1. Introduction
In the eastern countries, bladder cancer is the fourth most common cancer in men and the ninth most common in women, approximately 90% of malignant tumors arising in urinary bladder are of epithelial origin [1], however, it is the first most common in urinary and genitor system of Chinese men, and there is a increasing pattern throughout the world. Growth factor and their receptors regulate normal cell proliferation and differentiation and may be involved in any of the several steps in neoplastic development and progression. In several tumour types, including bladder cancers, over expression of EGFR has been shown to be correlated with poor prognosis [2-5].
The nuclear matrix is the framework or scaffolding of the nucleus and consists of the peripheral lamina, pore complexes, internal ribonucleic protein network, and residual nucleoli [6]. The importance of nuclear matrix is that they appear to provide spatial order and DNA domains for individual chromosome domains within the interphase nucleus. They also form fixed loci in the nucleus for individual sites of DNA replication, RNA transcription, and message RNA splicing, and steroid hormone nuclear regulation [7-12] and thus account for the differential nuclear matrix proteins (NMPs) expression between normal and cancer cells [7]. Some cancer specific NMPs were found in human cancers including bladder transitional cell carcinoma [13-19]. The relation between EGFR and nuclear matrix in human bladder cancer has not previously been studied.
In this work, we analyze the association of EGFR gene fragments with nuclear matrix in human bladder carcinoma, the binding potential of EGFR gene to the nuclear matrix proteins of bladder cancer with EGFR gene different gene loci was also studied.
2. Materials and Methods
2.1 Patients
Human bladder cancer tissue specimens were obtained from 15 men and one woman (stage I, 4 cases; stage II, 9 cases; stage III, 3 cases) who underwent surgery removal for cancer at the first and second hospital of Shantou University Medical College, China. The median age of the patients was 59.3 years (range: 19 to 79).
2.2 Tissue preparation
Fresh tissues were procured within 30 minutes of surgical removal of bladder cancer tissues. About 1 gram of tumor tissue was removed from 16 bladder cancer tissues. The specimens were trimmed to obtain 100 percent purity as far as possible and confirmed by histological diagnosis, The specimens were snap frozen and stored at -70°C.
2.3 Cell Culture
Human bladder transitional cell carcinoma cell lines T-24 were purchased from Shanghai Institute of Cell Biology, Chinese Academy of Sciences. T24 cells were grown in RPMI 1640 media (GIBCO-BRL), containing 10% heat-inactivated fetal calf serum (FCS), and 10,000U/ml penicillin G and 10mg/ml streptomycin sulfate. Cells were grown as adherent cells in a humidified atmosphere at 37°C in 5% CO2. Cells were digested by 2.5g/L trypsin and washed with cool PBS and collected in a new tube.
2.4 Nuclear matrix proteins extraction
NMPs were extracted by some modifications of the method described previously [20]. Briefly, the bladder cancer tissue specimens were minced into small (1-mm3) pieces, then these pieces or T24 cells were homogenized with a Tefl , and treated with a buffer�� solution including 0.5% NP-40, 0.25M sucrose, 100mM NaCI, 50mM KCI, 20mM Tris-CI, pH7.5, 10mM EDTA, 1mM phenylmethylsulfonyl fluoride (PMSF), to remove lipids and soluble proteins. The extracts were filtered through a 350mm nylon mesh to remove connective tissue and large debris, followed by a centrifugation step and collected supernatants (cytoplasm part), washed pellets two times by LS buffer including 10mM Tris-CI, pH7.5, 0.2mM MgCI2, 1mM PMSF. 2M sucrose cushion was prepared and paved the pellets onto it to purify nucleolus. Chromatin was removed by incubating the purify nucleolus pellets in DNAse I (100mg/ml, Worthington) in a buffered solution containing 1mM PMSF for 30 min at room temperature and ended digestion on ice, followed by a centrifugation step and washed two times by LS buffer. Soluble cytoskeletal proteins were removed by incubating and gently homogenizing the pellets in an HS buffer containing 10mM Tris-CI, pH7.5, 2M NaCI, 0.02mM DTT, 1.0mM PMSF for 30 min on ice, followed by centrifugation and washed in buffer I. The remaining pellet fraction, which contained nuclear matrix proteins and intermediate filaments, was solubilized in a disassembly buffer containing 8M urea, 1mM PMSF and 1%(v/v) 2-mercaptoethanol. Insoluble contaminants, primarily carbohydrates and extracellular matrix, were removed by ultracentrifugation. Intermediate filaments were allowed to reassemble upon removal of urea by dialysis in an assembly buffer containing 150mM KCl, 25mM imidazole HCl, 5mM MgCl2, 0.125mM EDTA, 2mM DTT and 1mM PMSF and removed by ultracentrifugation, leaving the nuclear matrix proteins in the supernatant fraction. Protein concentration of nuclear matrix proteins was determined by the Bradford method using BSA as standard.
2.5 Total cell protein extraction
Bladder cancer specimens were minced into small pieces and homogenized with Triple-detergent lysis buffer containing 50mM Tris-CI, pH8.0, 150mM NaCI, 100mg/ml PMSF, 0.1% SDS, 1% NP-40 and 0.5% sodium deoxychorate for one hour on ice. Debris was precipitated by centrifugation for 30 min at 16000g at 4°C. The supernatants of total cell extraction were stored at -70°C. Total cell proteins of T24 cells were performed as the above methods of Bladder cancer specimens.
2.6 Genomic DNA extraction
Freeze tissues or cultural T-24 cells were homogenated in digestion buffer; add 30μl of Proteinase K (stock solution of 20 mg/ml); Incubate at 50°C for overnight; the DNA from the lysate was then extracted by phenol-chloroform method and RNA was removed by DNase-free Rnase-A treatment at 37°C for one hour. After re-extracting with Phenol /Chloroform/ Isoamylalcohol. DNA was precipitated with ethanol in the presence of sodium acetate and stored in TE buffer at 4°C. DNA concentration was determined by a nuclear acid and protein analyzer DU640 (Beckman).
2.7 Nuclear matrix-associated DNA extraction [21-22]
Briefly, pure cellular nucleus of bladder cancer tissue specimens were obtained by the above method of NMPs extraction and separated into four equal parts. Chromatin was digested by incubating the pellets in DNase I (0, 25, 50, and 100mg/ml, Worthington) in a buffered solution containing 1mM PMSF for 30 min at room temperature respectively and ended reaction on ice, followed by a centrifugation step and washed two times in LS buffer. Then incubating and gently homogenizing the pellets in an HS buffer containing 10mM Tris-CI pH7.5, 2M NaCI, 0.02mM DTT, 1.0mM PMSF for 30 min on ice, followed by centrifugation, collected supernatants and pellets and re-extracted DNA by the method of genomic DNA extraction. They were named by H0DNA, H25DNA, H50DNA, H100DNA, and sDNA respectively.
2.8 Polymerase chain reaction
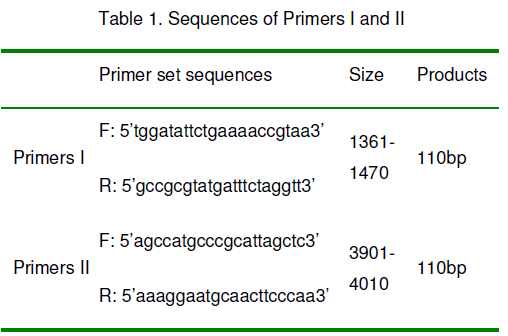
Two pairs primers were designed and synthesized according to the EGFR gene cDNA sequence with primers I at positions 1361-1470nt and primers II at positions 3901-4010nt [21-22]. These primers are listed on Table 1. Possible PCR products are 110bp.
The reaction was performed in 50ml volume of PCR buffer with 1.5mM MgCl2, 4��dNTPs and sample DNA. The two amplification reactions were performed in Mastercycle (eppendorf) with 32 cycles. Pre-denaturated at 95°C for 5 mins, denaturated at 95°C for 30 seconds, annealed at 58°C for 40 seconds, extended at 72°C for 1min and at last at 72°C for 7 mins (extension). The PCR products were finally electrophoresed on a 1.8% agarose gel in the presence of DNA ladders.
2.9 Probe preparation
Changes of protective adaptation can also reflect the effects of VAM on host plants. Soluble carbohydrate accumulation, amino and imino acids and protective enzyme activity are typically biochemical parameters used in studies on plant protective adaptation. Investigators have usually suggested that the differences of these biochemical parameters between VAM and NM plants reflect greater drought resistance in the VAM plants. For example, lower accumulation of soluble sugars indicates that plants avoid drought more successfully and so have less need to osmotically adjust symplasm or osmoprotect enzymes, or shows less strain or injury.
The probe was a PCR-amplified (primers II) 110bp fragment labeled by the DIG-High Prime DNA Labeling and Detection Kit (Roche), Labeling and detection assays were done using a nonradioactive method according to the manufacturer’s protocol.
2.10 Southwestern blot analysis [23]
50mg proteins (NMPs, total cell proteins) from human bladder carcinoma tissues or T24 culture cells separated by SDS-PAGE and transferred to nitrocellulose membrane (Sigma) in 50mM Tris, 40mM glycine, 20% methanol buffer using a transblot apparatus system (mini-protein III, Bio- Rad). The membrane was stained with Ponceau S (Sigma) and destained with de-ionized water; membrane-bound proteins were allowed to renature and block overnight at 4°C in TNED buffer (50mM Tris-HCl, pH7.5, 50mM NaCl, 0.1mM EDTA, and 1mM dithiothreitol) containing 5% non-fat milk powder, blots were washed with TNED buffer and then hybridized for overnight at room temperature using TNED buffer containing 10ng/ml of single stranded end labeled probes prepared by PCR product (primers II). After hybridization, the membrane was washed three times (10 mins each) with the TNED buffer, dried and subjected to detect by DAB kit.
3. Results
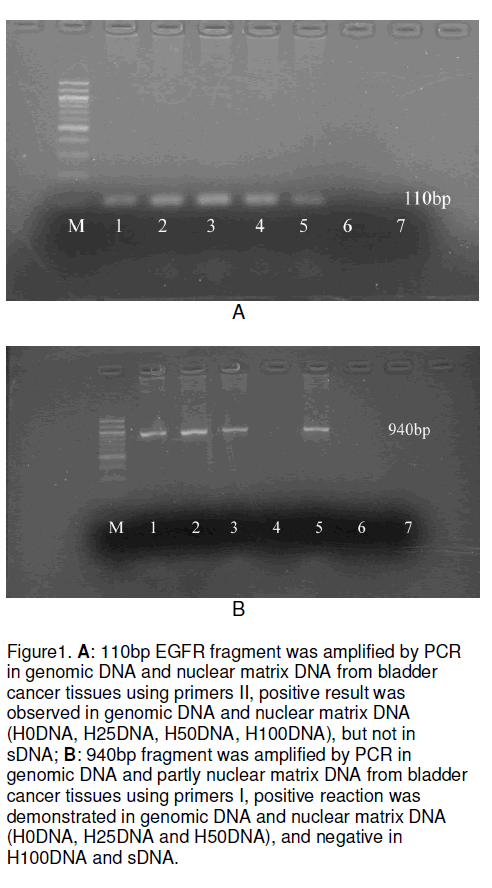
In order to identify binding of EGFR gene fragments to nuclear matrix proteins in bladder cancer, at first we use polymerase chain reaction to detect different fragment amplification of EGFR gene by ingredient nuclease digested DNA template, and discovered both genomic DNA and nuclear matrix DNA (H0DNA, H25DNA, H50DNA, H100DNA) of human bladder carcinoma could amplify 110bp positive fragment with primers II even the DNase I concentration was up to 100mg/ml, and it was negative in sDNA (Figure 1A). However both genomic DNA and nuclear matrix DNA (H0DNA, H25DNA and H50DNA) of human bladder carcinoma showed 940bp positive fragment with primers I, and not in H100DNA and sDNA (Figure 1B).
Figure 1:A: 110bp EGFR fragment was amplified by PCR in genomic DNA and nuclear matrix DNA from bladder cancer tissues using primers II, positive result was observed in genomic DNA and nuclear matrix DNA (H0DNA, H25DNA, H50DNA, H100DNA), but not in sDNA; B: 940bp fragment was amplified by PCR in genomic DNA and partly nuclear matrix DNA from bladder cancer tissues using primers I, positive reaction was demonstrated in genomic DNA and nuclear matrix DNA (H0DNA, H25DNA and H50DNA), and negative in H100DNA and sDNA.
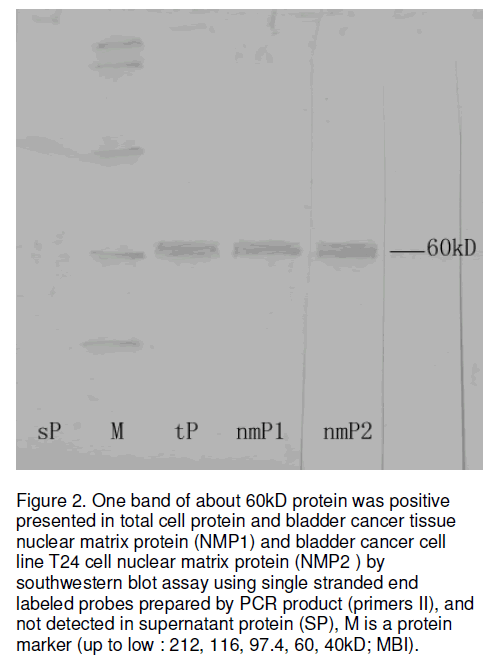
Secondly, we use Southwestern blot to further analysis the ability of binding of EGFR gene fragments to nuclear matrix proteins in bladder cancer and demonstrated that EGFR gene fragment (3901-4010nt) was bound to a about 60kD nuclear matrix protein in both NMPs and total cell protein samples, and not in cytoplasm sample (Figure 2).
Figure 2:One band of about 60kD protein was positive presented in total cell protein and bladder cancer tissue nuclear matrix protein (NMP1) and bladder cancer cell line T24 cell nuclear matrix protein (NMP2 ) by southwestern blot assay using single stranded end labeled probes prepared by PCR product (primers II), and not detected in supernatant protein (SP), M is a protein marker (up to low : 212, 116, 97.4, 60, 40kD; MBI).
4 Discussions
The epidermal growth factor receptor (EGFR) is a member of the tyrosine kinase receptor family; EGFR is the protein product of the c-erbB-1 protooncogene and is a 170kDa protein. The EGFR is proposed to participate in the pathogenesis or maintenance of a number of human cancers of epithelial origin and likely participation of the EGFR in epithelial carcinogenesis. EGFR is commonly over expressed in a number of epithelial malignancies and is often associated with an overall poor prognosis in patients with bladder cancer, previous reports have shown EGFR expression in 30-50% of bladder tumors [3, 24-25].
Key cellular processes such as DNA replication, transcription, and RNA processing and transport have been shown to be associated to nuclear matrix, nuclear matrix proteins serve as an important actively protein group to regulate carcinoma formation, cell cycle and cell proliferation, evidence of cell and tissue specificity of nuclear matrix proteins has been found. Tumor associated nuclear matrix proteins have also been identified in cancers of breast, colon, bone, larynx, kidney, prostate, liver, leukemia, cervix [13-19]. We previously found that hepatoma-assosiated nuclear matrix proteins are present in active chromatin too [20].
Wanng Z.H. et al [22] demonstrated that the binding property of the EGFR gene fragment to nuclear matrix is associated with metastatic potential of metastatic melanoma; EGFR-antisense may reduce tumorigenicity on human glioblastoma cells by changing nuclear matrix proteins compositions [26], so nuclear matrix proteins may closely associated with the effects of EGFR gene in the tumorigeness process.
In our study, both genomic DNA and nuclear matrix DNA (H0DNA, H25DNA, H50DNA, H100DNA) showed 110bp positive amplification patterns for the primers II (showed in Figure 1A). However, a 940bp specific band was detected in genomic DNA and nuclear matrix DNA (H0DNA, H25DNA, H50DNA) with primers I, not showed in H100DNA (showed in Figure 1B). These results suggest that EGFR genomic DNA fragments are associated with the nuclear matrix and that the nuclear matrix may involve in EGFR gene transcription and other gene regulation things. Even the digestion of DNase I concentration was up to 100mg/ml, a specific 110bp band was detected strongly in PCR products with primers II, and the positive 940bp band disappeared with primers I. This suggests that the binding of the EGFR gene fragment at position 1361-1470nt to the nuclear matrix is stronger than that at position 3901-4010nt in the human bladder transitional cell carcinoma. Because 940bp PCR product with primers I is surpass 110bp possible PCR product and then we search the 940bp fragment in Genbank but not discovered any record of its function. This results demonstrates that there maybe present an intron or gene regulated sequence between the two band primer (primers I) and strongly suggests that active EGFR gene fragment strongly associate with nuclear matrix in the human bladder transitional cell carcinoma, the detailed construction and function of this 940bp fragment maybe deserve to further study. On the other hand, our Southwestern blot analysis found that 110bp EGFR gene fragment was bound to a 60kD nuclear matrix protein strongly, this demonstrate that there is association between EGFR gene and nuclear matrix proteins in the human bladder transitional cell carcinoma.
4 Conclusion
Active EGFR gene is strongly bound to the nuclear matrix and nuclear matrix proteins, nuclear matrix proteins maybe anticipate in the high expression of EGFR gene including the course of transduction and post-transduction and/or translation in the human bladder transitional cell carcinoma.
Acknowledgements
This work was supported by the Medical Science Research Fund of Guangdong Province, China (grant No. 19983911).
References
- Greenlee R.T., Murray T., Bolden S., et al. (2000) Cancer Statistics 2000. CA Cancer J. Clin. 50: 7-33.
- Scagliotti G.V., Selvaggi G., Novello S., et al. (2004) The biology of epidermal growth factor receptor in lung cancer. Clin Cancer Res., 10(12): 4227-4232.
- Rajjayabun P.H., Keegan P.E., Lunec J., et al. (2005) erbB receptor expression patterns in human bladder cancer. Urology, 66(1): 196-200.
- Abd El-Rehim D.M., Pinder S.E., Paish C.E., et al. (2004) Expression and co-expression of the members of the epidermal growth factor receptor (EGFR) family in invasive breast carcinoma. Br J Cancer, 91(8): 1532-1542.
- Sjakste N., Sjakste T., Vikmanis U. (2004) Role of the nuclear matrix proteins in malignant transformation and cancer diagnosis. Exp Oncol, 26(3): 170-178.
- Berezney R. and Coffey D.S. (1974) Identification of a nuclear protein matrix. Biochem. Biophys. Res. Commun, 60: 1410–1417.
- Fey E.G., Bangs P., Sparks C., et al. (1991) The nuclear matrix: defining structural and functional roles. Crit. Rev. Eukaryotic Gene Expression, 1: 127-144.
- Pienta K.J., Partin A.W., Coffey D.S. (1989) Cancer as a disease of DNA organization and dynamic cell structure. Cancer Res, 49: 2525– 2532.
- Getzenberg R.H. (1994) The nuclear matrix and the regulation of gene expression: tissue specificity. J. Cell Biochem, 55: 22–31.
- Nelson W.G., Pienta K.J., Barrack E.R., et al. (1986) The role of the nuclear matrix in the organization and function of DNA. Annu Rev Biophys Biophys Chem, 15: 457-475.
- Pardoll D.M., Vogelstein B., Coffey D.S. (1980) A fixed site of DNA replication in eucaryotic cells. Cell, 19(2): 527-536.
- Getzenberg R.H., Pienta K.J., Coffey D.S. (1990) The tissue matrix: cell dynamics and hormone action. Endocr Rev, 11(3): 399-341.
- Konety B.R., Nangia A.K., Nguyen T.S., et al. (1998) Identification of nuclear matrix protein alterations associated with renal cell carcinoma. J. Urol, 159: 1359–1363.
- Getzenberg R.H., Konety B.R., Oeler T.A., et al. (1996) Bladder cancer-associated nuclear matrix proteins. Cancer. Res., 56: 1690–1694.
- Getzenberg R.H., Pienta K.J., Huang E.Y., et al. (1991) Identification of nuclear matrix proteins in the cancer and normal rat prostate. Cancer Res., 51: 6514–6520.
- Hughes J.H. and Cohen M.B. (1999) Nuclear matrix proteins and their potential applications to diagnostic pathology. Am. J. Clin. Pathol, 111: 267–274.
- Konety B.R., Nguyen T.S., Dhir R., et al. (2000) Detection of bladder cancer using a novel nuclear matrix protein BLCA-4. Clin. Cancer Res., 6: 2618- 2625.
- Lakshmanan Y., Subong E.N., Partin A.W., et al. (1998) Differential nuclear matrix protein expression in prostate cancers: correlation with pathologic stage. J Urol, 159(4): 1354-1358.
- Keesee S.K., Marchese J., Meneses A., et al. (1998) Human cervical cancer-associated nuclear matrix proteins. Exp Cell Res, 244(1): 14-25.
- Wen B.G., Burklard A.L., Cote G.J., et al. (1987) Hepatoma-assosiated nonhistone chromosomal proteins are present in active chromatin. Mol Cell Biochem, 76(1): 75-84.
- Konety B.R., Nguyen T.S., Brenes G., et al. (1984) Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. Nature, 309(5967): 418-425.
- Wang Z.H., Ding M.X., Jin M.L., et al. (1999) Association of EGFR gene fragments with nuclear matrix proteins in high and low metastatic melanoma cell lines. Anticancer Res., 19(5B): 4349-4352.
- Han X., Boyd P.J., Colgan S., et al. (2003) Transcriptional up-regulation of endothelial cell matrix metalloproteinase-2 in response to extracellular cues involves GATA-2. J Biol Chem., 278(48): 47785-47791.
- Sriplakich S., Jahnson S., Karlsson M.G. (1999) Epidermal growth factor receptor expression: predictive value for the outcome after cystectomy for bladder cancer. BJU Int, 83(4): 498-503.
- Gullick W.J. (1991) Prevalence of aberrant expression of the epidermal growth factor receptor in human cancers. Br Med Bull., 47(1): 87-98.
- Wang Z.H., Yam H.F., Tian X.X., et al. (1998) Comparison of nuclear matrix proteins between EGFRantisense transfected and untransfected glioblastoma cells. Inter J Mol Med, 2: 217-220.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences