Assessment of Molecular Variation in Pea Germplasm Using RAPD Markers
Zahra Alizadeh, Daryoosh Nabati Ahmadi, Mohtasham Mohammadi, Rahmat Allah Karimizadeh, H Rajabi-Memari, Moslem Bahmankar
Zahra Alizadeh1*, Daryoosh Nabati Ahmadi1, Mohtasham Mohammadi2, Rahmat Allah Karimizadeh2, H Rajabi-Memari1, Moslem Bahmankar3
1 Agronomy and Plant Breeding Department, College of Agriculture, Shahid Chamran University, Ahvaz, Iran
2 Dryland Agricultural Research Institute, Agricultural Research, Education and Extension Organization, Gachsaran, Iran
3 Department of Agronomy and Plant Breeding, College of Aburaihan, University of Tehran, Tehran, Iran.
- Corresponding Author:
- Zahra Alizadeh
Agronomy and Plant Breeding Department
College of Agriculture, Shahid Chamran University, Ahvaz, Iran
Tel: 989366379582
E-mail: alizadeh.scu.2012@gmail.com
Received date: October 22, 2015 Accepted date: December 08, 2015 Published date: January 14, 2016
Citation: Alizadeh Z, Ahmadi Nabati DA, Memari HR, et al., Assessment of Molecular Variation in Pea Germplasm Using RAPD Markers. Electronic J Biol, 12:1
Abstract
Pea (Pisum sativum L.) is an economically important pulse crop that grown for its protein-rich seeds. The Molecular Variation of 20 Pea genotypes supplied from different geographical regions of Iran and ICARDA was evaluated. 20 Random Amplified Polymorphic DNA (RAPD) primers were used in this study. 12 primers could generate 70 repeatable and detectable bands which 88% of them were polymorphic among various genotypes. Polymorphic information content (PIC) values ranged from 0 to 1 with an average of 0.82. The Jacard similarity coefficients among the genotypes ranged from 0.4 (between FLIP 01-32C and FLIP 93-93C) to 0.91 (between FLIP 03-71C and FLIP 03-64C) that indicate a relatively high genetic variation among the pea genotypes evaluated. Both cluster and Principal components analysis (PCA) clearly separated pea genotypes into two groups. The result showed that Iranian genotypes (Azad and Arman) were placed in first cluster in which its other members supplied from ICARDA; therefore, likely they have a common origin. This finding suggested that RAPD markers could be estimated genetic relationships and differences of pea germplasm.
Keyword
Cluster analysis; Molecular variation; Pea; RAPD marker.
1. Introduction
Pea (Pisum sativum L.) is an economically important pulse crop that grown for its protein-rich seeds. Pea belongs to the Leguminosae family, which has an important ecological advantage because it contributes to the development of low-input farming systems by fixing atmospheric nitrogen and it serves as a break crop which further minimizes the need for external inputs [1]. Pea is an important winter pulse crop in Western Europe, India, Australia, Pakistan, North and South America and is among the four important cultivated legumes after soybean, groundnut and beans [2,3]. Molecular variation has an important role in plant breeding programs. To evaluate genetic resources for their productivity, quality parameters and stress tolerance, field experiments are usually time consuming, therefore, molecular markers and DNA technology are used to assess diversity in the gene pool to identify genes of interest and to develop a set of markers for screening progeny [4-6]. The first step of any breeding program is assessment of Molecular Variation based on morphological characters and molecular data. Although sometimes morphological characters are useful, they are may be affected by environmental condition. Molecular markers are useful to complement the morphological and phonological characters because they are plentiful, independent of tissue or environmental effects and allow genotype identification in the early stages of development [2,7]. Random Amplified Polymorphic DNA (RAPD) is one of these Molecular markers that multi locus with the simplest and fastest detection technology [5]. RAPD have been successfully employed for assessment of Molecular Variation in several grain legumes. These include Lens sp. [8], Phaseolus sp. [9,10], Pea [11-13]. The objective of this study was to discern Molecular Variation of 20 genotypes of the pea with native and exotic origin using RAPD markers and its application to germplasm identification and classification.
2. Materials and Method
2.1 Plant materials and DNA extraction
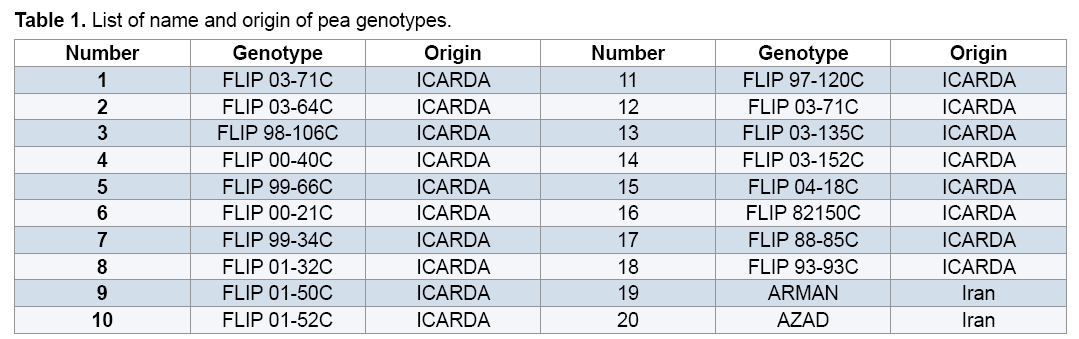
The study was performed on 20 pea genotypes which were supplied from Iran (2 genotypes) and ICARDA separate characters and scored for the presence (1) (18 genotypes) (Table 1).Total genomic DNA was extracted separately from young leaves of all the genotypes using by modifying the CTAB method [14]. After successful Extraction of genomic DNA, its quality and quantity was checked on 1% agarose gel and spectrophotometer and the samples were diluted to a final concentration of 50 ng/μl. Extracted DNA samples were stored at 4°C for Polymerase Chain Reaction (PCR) use.
2.2 DNA amplification and data analysis
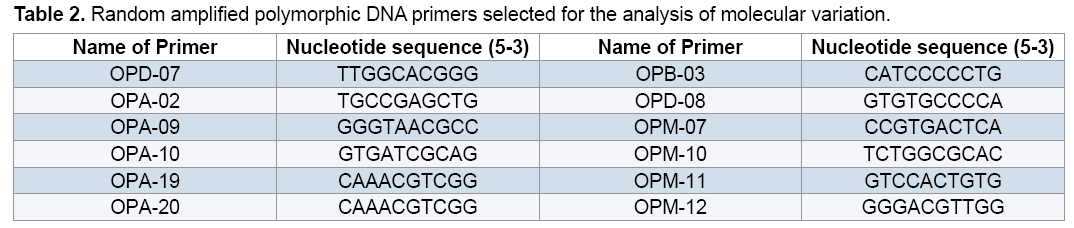
Out of 20 random 10-bp oligonucleotide primers used in this study 12 primers that showed amplified clear and reproducible band profiles were selected (Table 2). Components of each 25 μl PCR optimized or absence (0) of bands. The binary matrices were subjected to statistical analyses using NTSYS 2.02 [15]. Obtained the similarity matrix with NTSYS 2.02 were then used to cluster the data using Unweighted Pair Group Method with Arithmetic Mean (UPGMA) algorithm and Principal Component Analysis (PCA). To evaluate diverse level of each RAPD marker, Polymorphism Information Content (PIC) value of each RAPD marker was calculated using the formula: PIC=1- Σpi 2, where pi is the allele frequency of the ith allele [16].
3. Results and Discussion
3.1 Primers analysis
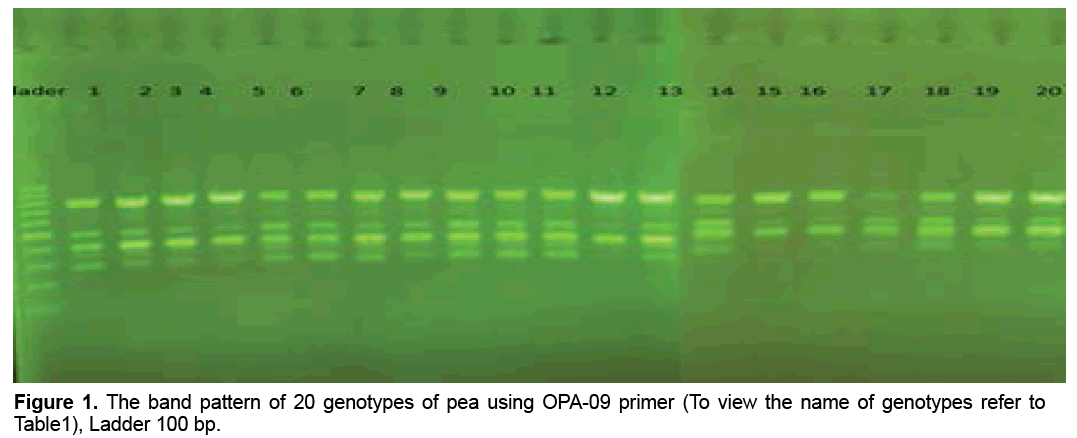
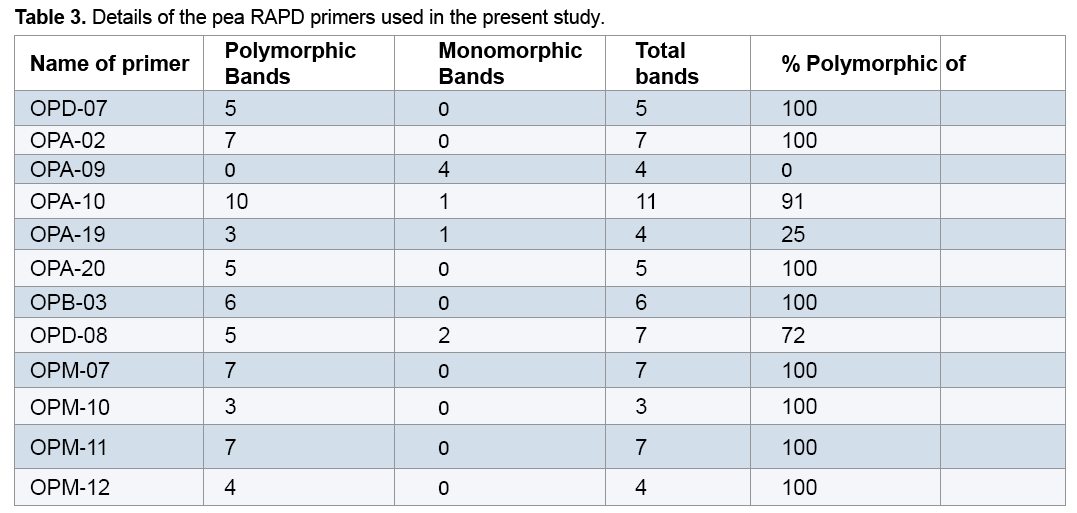
reaction mixture were including 15.95 μl Water distillated twice, 2.5 μl Buffer PCR (10x), 0.75 μl MgCl2 (50 mil mol), 2.5 μl Nucleotides mixture (2.5 mi molar), 1 μl Primer (10 pmol), 0.3 μl Tag polymerase and 2 μl DNA (50 ng). Amplification of DNA reactions were conducted in a DNA Thermo cycler machine (BIORAD, Model 1148) with programmed as follows; one hot start cycle of 94ºC for 5 min, 45 cycles of 94ºC for 30 second (DNA denaturation), 45 cycles of 39ºC for 50 second (annealing), 45 cycle of 72ºC for 50second (DNA extension) and a final extension one cycle of 72ºC for 10 min. The PCR products were electrophoresed on 1% agarose gels stained in SYBER Green and visualized on a UV light transilluminator. Band profiles were manually scored on two independent occasions and compiled into a binary matrix. Positive amplification were treated as From 20 primers RAPD employed in this study just 12 of them generated good amplification and reproducible band profiles (Table 3). Based on the appearance of polymorphism and repeatability of the bands, 12 primers could generate 70 repeatable and detectable bands which 88% (62 bands) were polymorphic among various genotypes. Study the pattern of bands was showed scale of 0, 1 and 0/82 for minimum, maximum and mean percentage of polymorphism for all primers, respectively. The level of polymorphism ranged from 0 to 100 (Table 3). The most informative loci of this work were (OPD- 07, OPA-02, OPA -20, OPB-03, OPM-07, OPM-10, OPM-11 AND OPM-12) with a PIC value of 100% while the less informative one was OPA-09 with a PIC value of 0% (Figure 1). Botstein et al. [17] believes if the polymorphism information contents value is greater than 0.5 this corresponds to a very informative marker. PIC values between 0.5 and 0.25 correspond to informative marker, and a PIC value less than or equal to 0.25 reflects lesser informative marker [17]. Therefore, according to our result the primers used in this study, with the exception of (OPA- 09 and OPA-19), were highly informative markers. Level of polymorphism observed in this study were more than results of Walunjkar et al. that reported the level of polymorphism was ranging from 78 to 94% [4]. Actually, aspects of practical grounds, OPD- 07, OPA-02, OPA -20, OPB-03, OPM-07, OPM-10, OPM-11 and OPM-12 primers that were produced the highest polymorphism and revealed the greatest potential to distinguish polymorphic DNA segments, can be recommended for future analysis of the pea genome using RAPD markers.
3.2 Cluster analysis
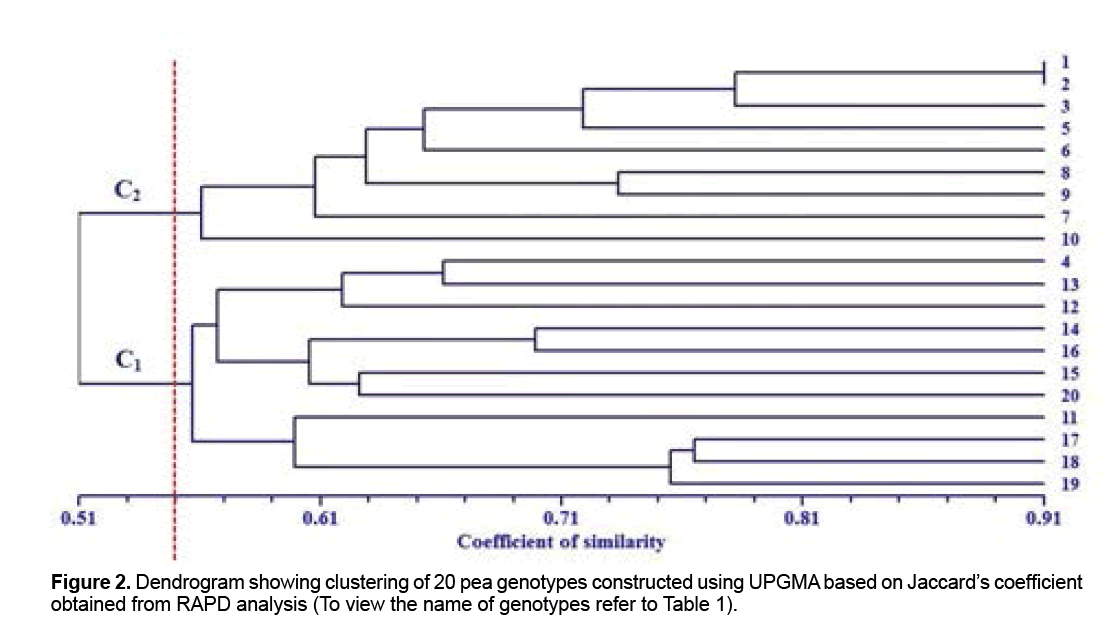
The cluster analysis based on molecular data in conjunction with Jacard similarity coefficient under UPGMA procedure was able to classify pea genotypes into two groups at the similarity level of 0.55 (Figure 2). The results showed that Molecular Variation is independent of origins of the genotypes. Iranian genotypes (AZAD and ARMAN) were placed in first cluster in which all its members supplied from ICARDA; therefore likely they have a common origin. Cluster 1 had the largest number of genotypes (11 genotypes) which originating from Iran (AZAD and ARMAN) and the remain genotypes supplied from ICARDA (Figure 2). Following first cluster, cluster 2 had the largest number of genotypes that all of them originated from ICARDA and including FLIP 03-71C, FLIP 03-64C, FLIP 98-106C, FLIP 99-66C, FLIP 00-21C, FLIP 99-34C, FLIP 01-32C, FLIP 01-50C and FLIP 01-52C. In this study, genotypes of FLIP 03-71C and FLIP 03-64C that clustered together in secondary group had highest degree of homology and similarity (0.91). According to this genetic similarity we could say that they probably have a common ancestor or could be an indicator of duplicates. However, similarities in genotypes can also increase due to convergent evolution, selection, or sharing of a common parentage [18]. Wani et al. observed similar results in a Molecular Variation study of five genotypes pea and classify them in two groups and suggested that the RAPD markers can be useful in breeding programs for assessment of the genetic similarity and as well as conservation of genetic resources [11]. In this study two genotype of FLIP 01-32C and FLIP 93-93C had lowest similarity coefficient (0.40) that implies they possess have different genetic backgrounds. Using information derived from RAPD markers, Plant breeders depends on genetic distance can select dissimilar genotypes as parental of crosses program pea to achievement maximize variability and more heterosis and transgressive segregation.
3.3 Principal component analysis (PCA)
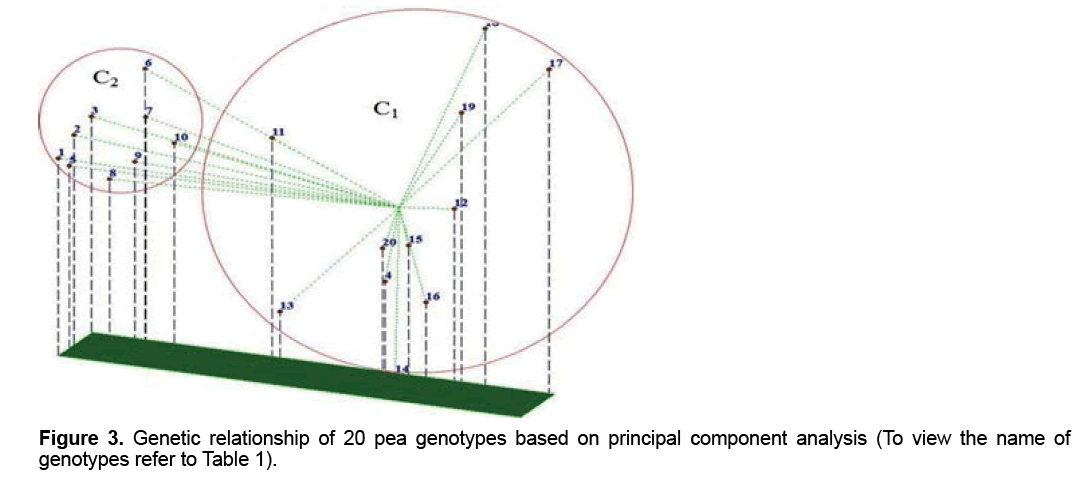
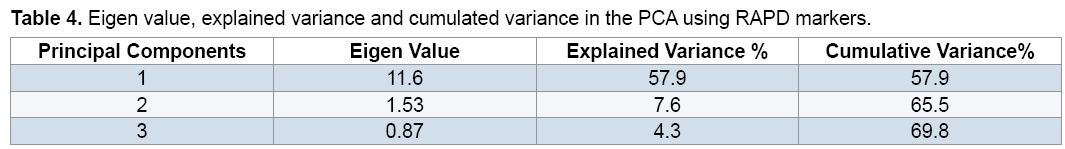
The PCA demonstrated that the first 3 components justified almost 69.8% of the total variation (Table 4). Although these results is suitable from perspective of statistical PCA, but according to genetic indicated unfavorable distribution of markers used in the genome and inappropriate sampling of them on the genome [19,20]. Principal component analysis based on molecular data precisely classified genotypes into two groups (Figure 3) and this result in consistent with result of cluster analysis. For instance, two genotypes, including FLIP 03-71C and FLIP 03-64C which were located together in the same group in cluster analysis, were also located close together in a diagram of principal component analysis. Therefore, these two methods of grouping, which were almost, similarly used separated genotypes from each other. Melchinger believes that if two or three of the first components of the principal component analysis justified small amount of the total variation (≤ 25%), principal component analysis encounter problems in genotypes clustering [20]. However, our results showed that the first three components justified almost 69.8% of the total variation; therefore, PCA method was usable to cluster genotypes and had no problem (Table 4).
4. Conclusion
The results of present study indicated that the RAPD markers can be useful in the Pea breeding programs. Molecular markers such as RAPD showed independent environment and plant growth stage; therefore, they can be employed to assess Molecular Variation. RAPD markers are useful for genotyping cultivars, but plant breeders to realize variation among germplasm still need the morphological data to complement molecular information. The result showed that Iranian genotypes (Azad and Arman) were placed in first cluster in which its other members supplied from ICARDA. The results of evaluation of molecular variation and far and near geographic distances of genotypes showed that generally geographic distance is not the reason for away and close genetics of individuals. Situate of genotypes related to different ecological conditions in the same cluster could be due to the existence of the identical genetic basis in them. This finding suggested that RAPD markers could be estimated genetic relationships and differences of pea germplasm.
References
- Smykal P, Aubert G, Burstin J, et al. (2012) Pea (Pisum sativum L.) in the Genomic Era. Agronomy. 2: 74-115.
- Bouhadida M, Srarfi F, Saadi I and Kharrat M. (2013). Molecular characterization of pea (Pisum sativum L.) using microsatellite markers. Journal of Applied Chemistry. 5: 57-61.
- Naghashzadeh, M, Beyranvand AF. (2015). Broad-Leaved Weeds in Chickpea (Cicer Arietinum L.) As Affected By Plant Density and Lentagran Herbicide Application.Electronic Journal of Biology. 11: 90-92.
- Walunjkar B, Parihar A, Chaurasia P, Pachchigar K, Chauhan RM. (2013).African Journal of Biotechnology. 12: 5823-5832.
- Karp A, Edwards K, Bruford M, et al. (1997). Newer molecular technologies for biodiversity evaluation: opportunities and challenges. Nature Biotechnology. 15: 625-628.
- Sun GL, William M, Liu J, Kasha KJ, Pauls KP. (2001). Microsatellite and RAPD polymorphisms in Ontario corn hybrids are related to the commercial sources and maturity ratings. Molecular Breeding. 7: 13-24.
- Taran B, Zhang C, Warkentin T, Tullu A, Vandenberg A. (2005). Molecular Variation among varieties and wild species accessions of pea (Pisum sativum L.) based on molecular markers, and morphological and physiological characters. Genome 48: 257-272.
- Sultana T, Ghafoor A. (2009). Botanical and molecular evidences of landraces from the germplasm exclusively collected from Baluchistan, a center of diversity for Lens culinaris. African Journal of Biotechnology. 8: 5310-5315.
- Johns MA, Skroch PW, Nienhuis J, et al. (1997). Gene pool classification of common bean landraces from Chile based on RAPD and morphological data. Crop Science. 37: 605-613.
- Chiorato AF, Carbonell SAM, Benchimol LL, et al. (2007). Molecular variation in common bean accessions evaluated by means of morpho-agronomical and RAPD data. Scientia Agricola (Piracicaba, Brazil). 64: 256-262.
- Wani GA, Mir BA, Shah MA. (2013). Evaluation of diversity in pea (Pisum sativum L) genotypes using agromorphological characters and RAPD analysis. International journal currency research review. 5: 17-25.
- Mulk khan S, Ahmad H, Nisar M, et al. (2013). Molecular variation in pea germplasm using Random Amplified Polymorphic DNA Markers. Pakistan Journal Botany. 45: 1259-1264.
- Choudhury PR, Singh IP, George B, Verma AK, Singh NP. (2008). Assessment of Molecular Variation of pigeon pea cultivars using RAPD analysis. Biological Plantarum. 52: 648-653.
- Ram G, Bhan MK, Gupta KK, Thaker B, Jamwal U and Pal S, 2005. Variability Pattern and Correlation Studies in Silybum Marianum Gaertn. Fitoterapia. 76: 143-147.
- Rohlf FJ, 1998. NTSYS-pc: Numerical taxonomy and multivariate analysis system, v. 2.0. Exeter Software. Setauket, New York.
- Anderson JA, Church JE, Autrique SD, Thanksley S, Sorrells ME. (1993). Optimizing parental selection for genetic linkage map. Journal of Genome. 36: 181-188.
- Botstein D, White RL, Skolnick M and Davis RW. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms 2: 314-331.
- Johnson RC, Kisha T, Foiles C and Bradley V. (2007). Characterizing safflower with AFLP Molecular Markers. Crop Science. 47: 1728-1736.
- Franco J, Crossa J, Villasenor J, Taba S and Eberhart SA. (1997). Classifying Mexican maize accessions using hierarchical and density search methods. Crop Science. 37: 972-980.
- Rohlf FJ. (1972). An empirical comparison of three ordination techniques in numerical taxonomy. Systematic Zoology. 21: 271-280.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences