Induction of Hydrolases and Phenylalanine Ammonia-lyase by Pathogen Derived Elicitors in Mungbean (Vigna radiata L.)
Deepak Koche, Arvind Choudhary
1Department of Botany, Shri Shivaji College, Akola (MS) India -444 005
2Dept. of Botany, RTM Nagpur University Campus, Nagpur (MS) India -440 033
- Corresponding Author:
- Email: dipakkoche43@gmail.com
Abstract
The enzyme activities of two hydrolases and phenylalanine ammonia-lyase were analyzed in the seedlings of four mungbean cultivars (TARM-1, Kopergaon, ML-936 and Pant M-3) differing in their resistant status. Nine day after germination, the seedlings were challenged with Cercospora canescens cell wall elicitor and spore suspension and they were evaluated periodically for the enzyme activities. The hydrolases i.e. chitinases and -1,3 glucanases were found to be induced in all seedling parts by both inducers. The activities of these enzymes were higher in resistant cultivars. The maximum activity of glucanase, chitinase and PAL was observed in cotyledons of ML-936 followed by Pant M-3. This induced activity of defense related enzymes was found to be associated with resistant status of mungbean cultivars.
Keywords
Vigna radiata, Cercospora canescens, chitinases, -1,3 glucanases, phenylalanine ammonia-lyase.
1 Introduction
Mungbean is among the major pulse crops. Though a number of varieties with different phytopathological profile were developed, its average yield remains low. It is mostly due to various fungal diseases. Cercospora leaf spot (CLS) is one of the most damaging foliar diseases that adversely affect the yield of mungbean cultivars. Therefore, to develop the variety with multiple diseases resistance is one of the major objectives in mungbean breeding program. In this context it is necessary to understand the resistance mechanism in this crop.
Hydrolases and phenylalanine ammonia-lyase (PAL) are actively involved in plant defense. The antifungal activity of hydrolases i.e. chitinases, -1,3 glucanases can be induced by exogenous application of different biotic and abiotic inducers have been demonstrated in several crop plants [1-4]. PAL is one of the key enzymes involved in the phenylpropanoid biosynthesis. The first reaction of the phenylpropanoid biosynthetic pathway is the deamination of phenylalanine to cinnamic acid by PAL which ultimately culminates in the synthesis of antimicrobial compounds. There are sufficient evidences supporting the induction of PAL by various biotic and abiotic stresses [5]. Such research in Mungbean-Cercospora pathosystem is still lacking. Therefore, this work is undertaken with the objectives to know the basics underlying the defense mechanism in above pathosystem and the role of hydrolase enzymes and PAL in mungbean defense system.
2 Methods
2.1 Procurement of Germplasm
The seeds of four mungbean cultivars used in this investigation, were procured from BARC, Mumbai (TARM-1 and Kopergaon), CPRI, Kanpur (Pant M-3) and Panjab Agricultural University, Ludhiana (ML- 936). The seeds were grown in the field and their resistance status was confirmed.
2.2 Elicitor Preparation
The Cercospora canescens was cultured on modified Czapek’s media supplemented with casein hydrlysate (20mg/l) and mungbean leaf extract (2gm/l). The 3-week old Cercospora canescens culture was used to prepare cell wall elicitor and spore suspension. The elicitor preparation was done as per Koche and Choudhary [6].
2.3 Elicitation Process
Nine-day old etiolated seedlings were used to elicit the defense response. The seedlings were aseptically sprayed with above concentration of pathogen derived elicitor. Distilled water was used as control. All the operations were performed in laminar air flow chamber. After elicitation, the seedlings were frozen in Liquid N2 every 5h up to 75h.
2.4 Enzyme analysis
The elicitor induced expression of glucanase, chitinase and PAL genes were analyzed biochemically by assaying the activity of these gene products (enzymes). Glucanase and chitinase we’re extracted by homogenizing 1gm of frozen tissue in 1ml ice cold extraction buffer (0.1M sodium citrate buffer, pH 5), in pre-chilled mortar and pestle. The extract was centrifuged at 10,000 rpm for 15 minutes. The 1ml of supernatant was taken in 1.5ml of centrifuge tube and proteins were precipitated by adding ammonium sulphate to saturation. Precipitated proteins were centrifuged at 10,000 rpm for 15 minutes and supernatant was discarded. The protein pellet was re-suspended in 1ml extraction buffer and used as enzyme source for glucanase and chitinase assay.
PAL was extracted from frozen tissue by homogenizing 1 gm tissue in 1 ml ice-cold extraction buffer, in pre chilled mortar and pestle. To avoid the interference in spectrophotometric readings, due to the presence of phenolics particularly cinnamic acid, 0.5% (w/v) Polyvenylpyrroledone (PVP) was added at the time of extraction to remove (adsorb) phenolics. The extract was centrifuged at 10,000 rpm and clear supernatant was used as enzyme source.
The glucanase assay was performed according to the procedure given by Kauffmann et al. [7]. The assay mixture containing 0.48 ml of 0.1M sodium acetate buffer (pH 5.2), 100ml enzyme extract and 200ml laminarin (5mg/ml) (Sigma make) was incubated at 37°C for 3h. Then 0.5ml alkaline copper tartarate was added to it and mixture was heated to 100°C for 5 minutes. The mixture was then cooled to room temperature and 0.5ml of arseno-molybdate reagent was added to it. After the development of blue color, 3ml of distilled water was added to each sample and absorbance was recorded at 660 nm against the blank containing the enzyme and all other reagents, except laminarin.
The chitinase assay was performed according to the process given by Boller et al. [2]. The assay mixture contained 100ml sodium acetate buffer (pH 4.5), 100ml sodium azide solution, 200ml of colloidal chitin (10mg/ml) (Himedia make) and 100ml of enzyme extract. The volume of this enzyme mixture was adjusted to 1ml by extraction buffer and incubated at 37°C for 3h. After incubation, 100ml of sodium borate buffer was added to the reaction mixture and heated to 100°C in water bath for 3 minutes. The mixture was then cooled in tap water and centrifuged at 10000 rpm for 5 minutes. The clear supernatant was collected and 3ml of DMAB reagent was added to it. The mixture was then incubated for 20 minutes at 37°C . The absorbance was recorded immediately at 585 nm against blank containing enzyme and other reagents except chitin.
For assaying PAL, two different sets of each sample were prepared. The vials of first set contained 100 ml enzyme extract and 2.9 ml of L-Phenylalanine (substrate) and second set contained 100 ml of enzyme extract and 2.9 ml of D-Phenylalanine solution (substrate check). The blank was prepared for each set separately. The blank of first set contained 100 ml L-Phenylalanine and 2.9 ml extraction buffer and of second set contained 100 ml D-Phenylalanine and 2.9 ml extraction buffer. These vials were then incubated in a water bath, at 40°C for 1 hour. Then absorbance was recorded at every 30 minutes interval upto 2 hours at 290 nm. All the assays were done in triplicate.
The protein content of each sample was estimated by using the method given by Bradford [8].
3 Results and Discussion:
The expression of two hydrolases i.e -1,3 glucanases and chitinases and PAL was investigated in terms of their enzyme activities induced by Cercospora cell wall elicitor and its spore suspesion, in different seedling parts of four mungbean cultivars differing in their resistance status. The induced activity of each hydrolases was then correlated with the resistant status of respective cultivar. The induction of chitinase and -1,3 glucanase was periodically studied in all seedling parts and cotyledons of four cultivars (two resistant and two susceptible).
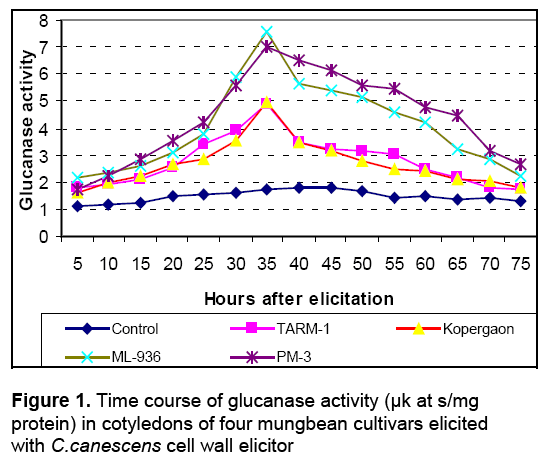
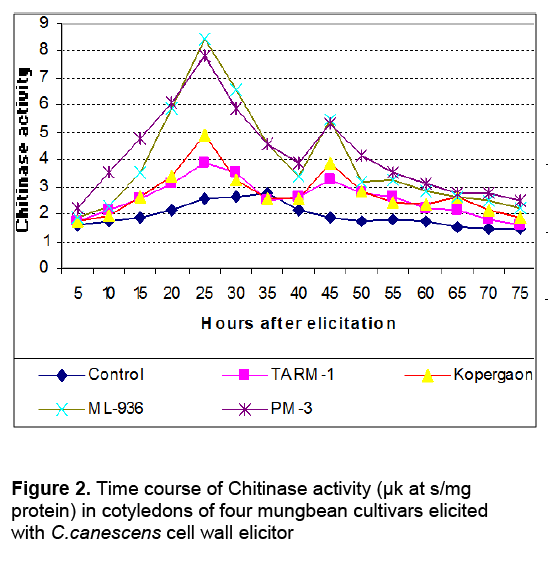
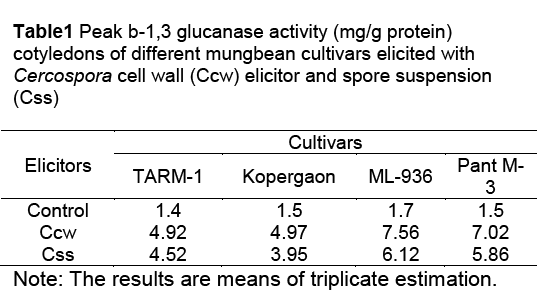
The activities of glucanases and chitinases were found to be induced as early as 5h after elicitation in cotyledons of all cultivars with Cercospora cell wall (Ccw) elicitor. The enzyme activities were gradually increased during post elicitation period and reached to its peak and then declined (Figure 1 and Figure 2). Throughout post elicitation period, the activities of glucanases and chitinases remained high in resistant cultivars as compared to susceptible ones. The glucanases and chitinases attained the peak at different periods. The glucanase showed only one peak of its high activity i.e. 35h after elicitation (Figure 1). However, chitinases displayed two peaks of higher activity. The first peak was observed after 25h and second was noticed 45h after elicitation (Figure 2). The similar pattern of induction of these enzymes was found in all other seedling parts of resistant and susceptible cultivars (Data not shown). The similar response is observed in all other seedling parts after elicitation with the C. canescens cell wall elicitor. The spore suspension treated seedling parts also revealed remarkable induction of hydrolases and show similar trends of accumulation (Table 1).
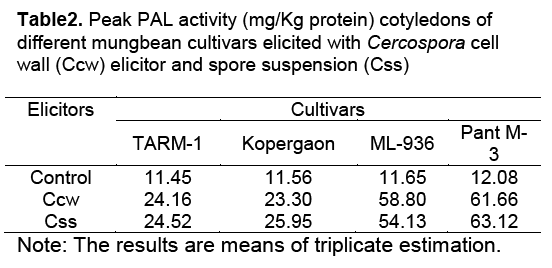
After challenging with both pathogen derived elicitors, PAL activity in seedling parts and cotyledons of both resistant and susceptible cultivars continue to increase till it reaches the peak, 8h after elicitation. But the peak activity in all seedling parts of resistant cultivars was found higher than in susceptible cultivars. The highest PAL activity was measured in elicited cotyledons of Pant M-3, followed by ML-936 (Table2).
In this investigation, resistant and susceptible cultivars displayed differential responses to Ccw elicitor and its spore suspension. In resistant cultivars the response was rapid and vigorous. The resistant cultivars showed 3.9 to 5 times more activity of these enzymes than in control (Table 1, Table 2 and Table 3). Whereas in the susceptible cultivars, the response was slow and activity was less as compared to resistant cultivars.
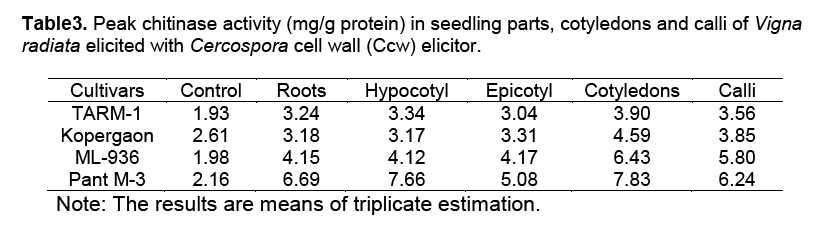
The hydrolases like -1,3 glucanases and chitinases were known to have antifungal activities and PAL synthesizing the antimicrobial compounds, thus are involved in conferring resistance to various plants [4,9-11]. The results of present investigation also substantiate this in mungbean-Cercospora canescens pathosystem. Both hydrolases and PAL were induced rapidly and their activity was significantly higher in all seedling parts of resistant cultivars (ML-936 & Pant M-3). Their induction in seedlings of susceptible cultivars (TARM-1 & Kopergaon) was slow and their peak activity was much less. Thus, the resistant mungbean cultivars showed hypersensitive response for the induction of hydrolases by Cercospora cell wall elicitor and UV-C.
The higher accumulation of hydrolases and PAL has been reported in resistant interactions of Maize, Pepper, Barley, Wheat and Pearl millet with pathogenic fungi [4,5,11-13]. These reports suggested that glucanases and chitinases play a vital role in determining the resistant status of plant against pathogens, either alone or in combination and PAL act as key enzymes in synthesis of antimicrobial compounds. In the present investigation it was found that both hydrolases and PAL are induced simultaneously and possibly synergistic action of hydrolases and high level of PAL leading antimicrobial compound synthesis defend the host from invading pathogen.
The induction studies of hydrolases and PAL suggest their involvement in determining the resistance in mungbean to Cercospora canescens. Differential expression of these enzymes in resistant and susceptible cultivars indicates the possible use of these enzymes as the biochemical markers for screening mungbean cultivars against Cercospora leaf spots (CLS) disease.
References
- Boller T. 1985: Induction of hydrolases as a defense reaction against pathogens, (In: Key J. L. Kosuge T eds.), Cellular, and Molecular biology of plant stress. New York, NY, USA: Alan, R Liss, 247- 262.
- Boller T. Gehri A. Mauch F. and Vogeli V. 1983: Chitinase in bean leaves: induction by ethylene, purification, properties and possible function. Planta 157: 22- 31.
- Mauch F. Mauch M. B. and Boller T. 1988: Antifungal hydrolases in pea tissue, II. Inhibition of fungal growth by combinations of chitinase and b-1,3 glucanase. Pl Physiol 88(3): 936- 942.
- Kim Y. J. and Hwang B. K. 1994: Differential accumulation of b-1,3 glucanase and chitinase isoforms in pepper strems infected by compatible and incompatible isolates of Phytophthora capsici. Physiol Mol Plant Path 45: 195- 209.
- Kale M. C. and Choudhary A. D. 2001: Induction of phenylalanine ammonia- lyase in groundnut cutltivars in response to biotic and abiotic stress Indian Phytopath. 54(3): 288-292.
- Koche D. K. and Choudhary A. D. 2005: Elicitor induced expression of b-1,3 glucanase and chitinase genes in Vigna radiata (L.) Wilczek Bionature 25 (1&2): 69-74.
- Kauffman S. Legrand M. Geoffroy P. and Fritig B. 1987: Biological function of Pathogenesis- related proteins: Four PR- proteins of tobacco have b-1,3 glucanase activity. EMBO J 6: 3209-3212.
- Bradford M. M. 1976: A rapid and sensitive method for quantification of microgram quantities of protein, utilizing the principal of protein dye binding. Anal Biochemistry. 72: 248-252.
- Van den Elzen P.J.M. Jongedijk K. Melchers L.S. and Cornelissen B.J.C. 1993: Virus and fungal resistance: from laboratory to field. Philosophical Transactions of Royal Society B 342: 271- 278.
- Van Loon L. C. 1997: Induced resistance in plants and the role of pathogenesis related proteins. Eur J Plant Pathology 103: 753- 765.
- Campos A. D. Ferreira A. G. Hampe M. M. V. Antunes I. F. Brancao N. Eilveira E. P. Da Silva J. B. and Osorio V. A. 2003: Induction of chalcone synthase and phenylalanine ammonia –lyase by salicylic acid and Colletotrichum lindmuthianum in common bean. Braz. J. Plant Physiol. 1(3): 129-134.
- Cordero M. J. Reventos D. and Segundo B. S. 1994: Expression of maize proteinase inhibitor gene is induced in response to wounding and fungal infection. Systemic wound- response to a monocot gene. Plant J. 6: 141- 150.
- Kini K.R. Vasanthi N. S. and Shetty H. S. 2000: Induction of b-1,3 glucanase in seedlings of pearl millet in response to infection by Sclerospora graminicola. Eur J Pl Physiol 106: 267- 274.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences