Homology model of 2C-methyl-d-erythritol 2, 4- cyclodiphosphate (MECP) synthase of Plasmodium falciparum 3D7
Neelima Arora, Amit Kumar Banerjee, U.S.N Murty
Bioinformatics Group,Biology Division,Indian Institute of Chemical Technology,Uppal Road,Tarnaka,Hyderabad-500007,India
- Corresponding Author:
- U.S.N Murty
Bioinformatics Group
Biology Division,Indian Institute of Chemical Technology
Uppal Road,Tarnaka,Hyderabad-500007,India
Tel: +91 040 27193134
Fax: +91 040 27193227
E-mail: murty_usn@yahoo.com
Abstract
Malaria has been a cause of enormous morbidity and mortality since the dawn of evolution. Control of malaria remains a farfetched dream despite numerous efforts and intervention strategies devised by research communities. Increasing drug resistance in the malarial parasite to conventional drugs has raised an alarm. This situation warrants the need for exploring novel drug targets. In this study, we report the structural modeling of an attractive drug target 2C-methyl-d-erythritol 2, 4- cyclodiphosphate (MECP) synthase. Threedimensional model of the enzyme was constructed using in-silico tools. The model was further evaluated for stereo-chemical quality. This model will provide an insight about the structure of MECP synthase and aid in rational drug design
Keywords
Homology modeling; Plasmodium falciparum; MECP synthase; molecular modeling; bioinformatics.
1. Introduction
Apicomplexa is a group of organisms characterized by the presence of four-membraned relict plastid [1],which is presumed to be acquired from eukaryotic red or green algae by secondary endosymbiosis during the course of evolution [1-7]. This fact has been established by identification of 35 KB extrachromosomal DNA in P. falciparum and about 551 nucleus-encoded apicoplast targeted (NEAT) proteins have been predicted [8],many of which are considered attractive drug targets [9]. This organelle is essential for survival of these parasites and houses many biochemical pathways like fatty acid,heme and isoprenoid biosynthesis [10]. These pathways are exclusively present in bacteria,plant and apicomplexan parasites but absent from human. This fact makes them attractive source of numerous drug targets [11-12]. Though isoprenoids may seem very diverse in structure and perform a variety of functions ranging from ETS,photosynthesis,signalling,control of biosynthesis of lipid,meiosis,apoptosis,protein degradation,the basis unit remains 5-carbon isoprene unit [13].
There are 2 distinct pathways to carry out isoprenoid biosynthesis [13],mevalonate pathway [14] and plastidial methylerythritol 4-phosphate (MEP) pathway/ Rohmer pathway [15-17]. The first pathway was presumed to be ubiquitous and the only route for synthesis of isoprenoid until recent discovery of mevalonate-independent MEP pathway in chloroplast of plants,eubacteria and apicomplexa. Plasmodium utilizes MEP pathway exclusively and this fact has been supported by studies involving inhibition of mevalonate pathway by mevastatin [18]. Several enzymes of this pathway have been identified including 1-deoxy-D-xylulose-5-phosphate synthase,1-deoxy-D-xylulose-5-phosphate reductoisomerase and 2C-methyl-D-erythritol 2,4- cyclodiphosphate synthase [13,19]. The increasing multi-drug resistance and spread of these Plasmodium strains are the cause of concern for public health authorities and failure of conventional drugs in controlling malaria has motivated researchers across the globe to explore new biochemical pathways in search of novel drug targets for developing efficient anti-malarials.
Owing to their absence in human hosts,this pathway represents an excellent source of novel drug targets and it is presumed that specific inhibitors designed against enzymes of this pathway will aid in designing new anti-infective agents,which will be less toxic and cause fewer side effects [20- 21]. MECPS,EC: 4.6.1.12) the fifth enzyme of MEP pathway catalyzes the formation of isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate [13] and represents a promising drug target for structure-based drug design. In the present study,we have attempted to construct the structure of MECP synthase using comparative modeling technique.
2. Materials and Methods
Retrieval of the sequence
Amino acid sequence of enzyme 2C-methyl-derythritol 2,4-cyclodiphosphate (MECP) synthase of Plasmodium falciparum 3D7 (Accession No: XP_001349603.1) was obtained from NCBI,a publicly available database. As experimentally determined structure of this enzyme is not available till date,we have attempted to determine its structure using in silico tools. Important properties of MECP synthase were calculated using PROTPARAM [22]. SOPMA [23] was employed to predict the secondary structure of the enzyme using 4 states prediction option and keeping a window width of 17 and similarity threshold of 8.
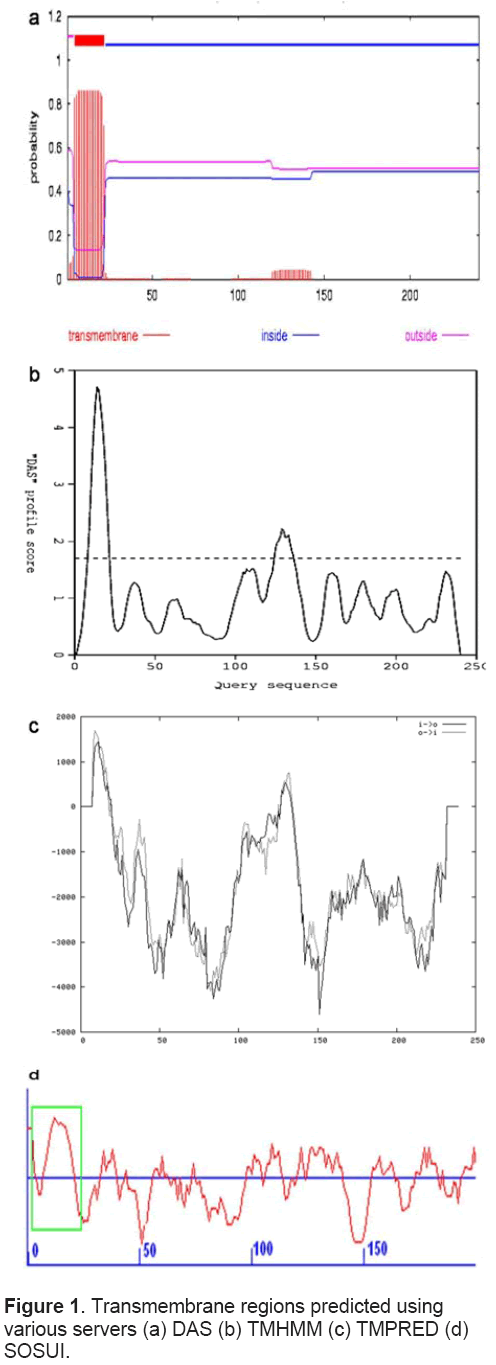
Identification of transmembrane regions
DAS [24],TMHMM [25],SOSUI server [26],TMPred [27] and HMMTOP [28] were employed to recognize the transmembrane regions in MECP synthase.
Selection of template
Template protein is selected based on sequence similarity. Template was searched by querying the target sequence against PDB database [29] using Protein-Basic Local Alignment Search Tool (BLASTp). Default parameters i.e. E-value threshold 10,word size 3 and Blosum 62 Matrix were considered for the same. Structure of 2C-methyl-derythritol 2,4-cyclodiphosphate of Plasmodium vivax (PDB ID: 3B6N_A) at a resolution of 2.26 Angstrom sharing 60% identity with target protein was selected for structural modelling.
Alignment of target-template
Amino acid sequence alignment of target and template proteins was derived using the ClustalX program [30] applying the default parameters. Target-template alignment is illustrated in Figure 2 . Default parameters were applied and the alignment was used as input for the modelling exercise.
Construction of 3-dimensional structure
MODELLER9v7 software [31],which exploits sequence alignment between the considered protein and its selected template,was used for determining tertiary model of the protein. This wellknown program performs a restraint based modelling,Complete assessment and evaluation of the generated models were performed followed by Ramachandran Plot analysis. Structure of the model was visualized using PYMOL [32] and VMD software [33].
Validation of the derived structure
For ascertaining the reliability of the obtained structure,model was subjected to evaluation of stereo-chemical quality using RAMPAGE [34].
Active site identification
Q-siteFinder [35] was employed for identification of binding sites in the derived structure.
3. Results and Discussion
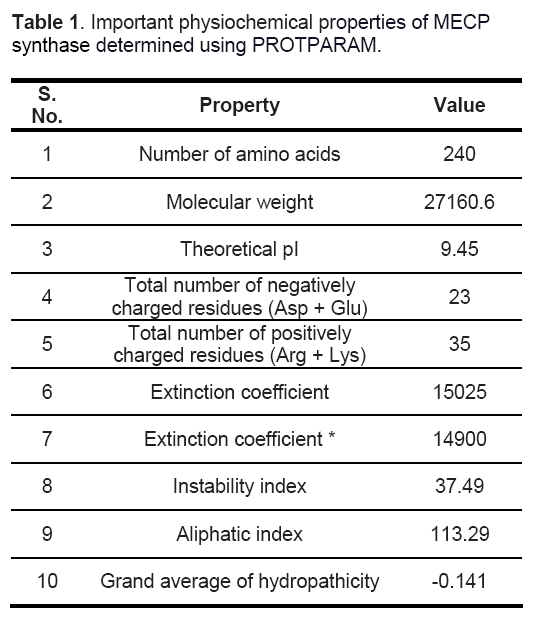
Important physiochemical properties of the target protein were computed using Protparam tool [22] to gain an insight about the protein (Table 1). MECP synthase is a basic protein having 240 residues. Instability index of 37.39 indicates the stable nature of protein and a low GRAVY value reflects its hydrated state.
Total 4 transmembrane regions (9-21,10-20,125- 136,129-129) were predicted in MECP synthase using DAS server but the range 129 -129 was ignored (Figure 1a) as it represents only 1 amino acid,TMHMM,SOSUI and HMMTOP servers also predicted occurrence of 1 transmembrane region spanning from 5-22,6-22,2-24 respectively (Figure 1b,1c and 1d). TMPRED predicted 2 inside to outside (2-22 and 120-139) and 2 outside to inside (1-21 and 120-143) helices. On comparison,most probable transmembrane regions showed concurrence as all tools predicted the transmembrane region spanning first 25 amino acids.
It was found that random coils (36.25%) are predominant among the secondary structure features followed by alpha helices (30.42%),extended strands (27.50%) and beta turns (5.83%).
In wake of a huge gap in the available protein sequences and number of structures deposited in PDB,researchers across the globe have adopted homology-modelling technique as a valid substitute to experimental techniques for structure determination. Comparative modelling is based on the fact that if two proteins share a good sequence identity,they also share structural similarity. This methodology has been utilized extensively for generation of 3D structure of proteins in case of Plasmodium also,for example apical membrane antigen [36],heat shock protein [37],Dihydrofolate reductase [38] and Thioredoxin reductase [39].
For obtaining target-template alignment,ClustalX using a gap penalty of 10 and a gap extension penalty of 0.05 was employed. Target-template alignment used for subsequent model generation is represented in Figure 2 .
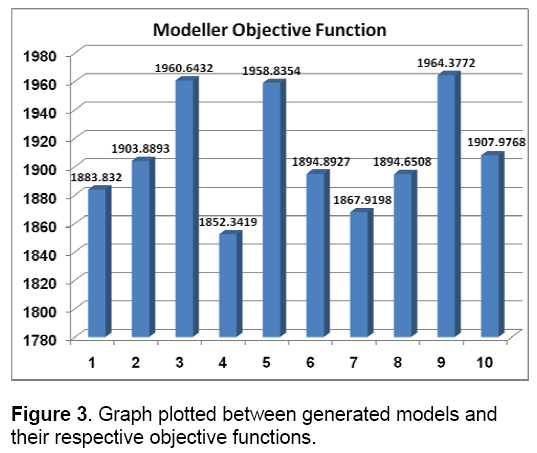
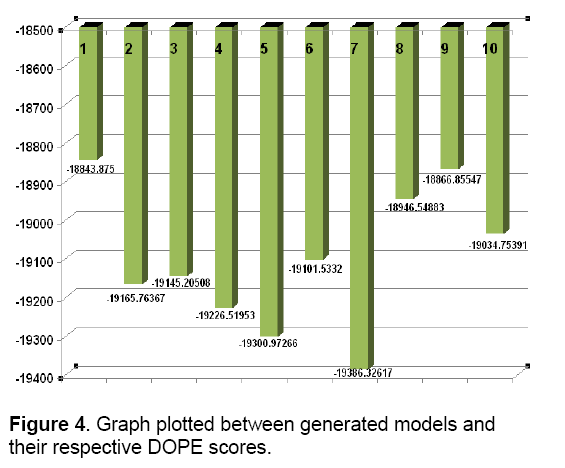
MODELLER relies on probability density functions as spatial restraints [31]. This approach resulted in 10 models. Modeller objective functions and DOPE (Discrete optimized potential energy) scores of the obtained models are shown in Figure 3 and Figure 4.
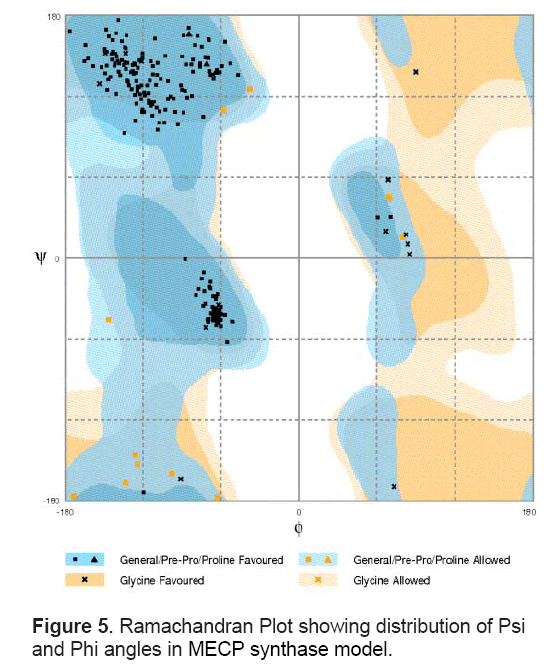
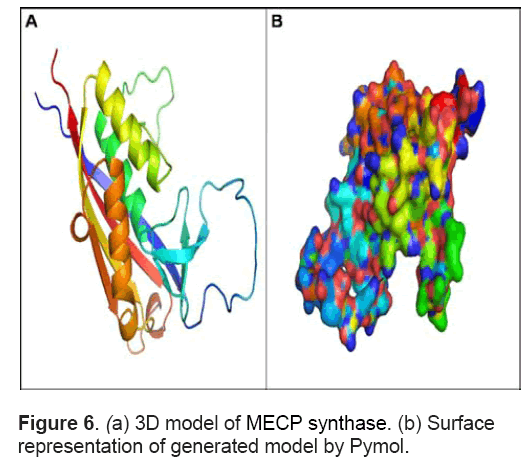
These models were assessed and evaluated using RAMPAGE server that considers dihedral angles ψ against φ of amino acid residues in protein structure. The model with no outliers showing 95% amino acids in favoured regions and 5% regions in allowed regions of Ramachandran plot was selected for further analysis (Figure 5 ). The structure has 3 large α helices and 4 large β sheets (Figure 6 A). Apart from these,it contains 4 small sheets. Out of four large β sheets,one pair is parallel whereas the other pair is anti -parallel in orientation. The large sheet in anti-parallel direction spanned from Ile 65 to Lys76 and the other ranged from Gly219 to Lys 238. The parallel sheets spanned from Aln174- Ile180 and Glu205-Thr214. Figure 6 B shows the surface representation of the model represented in default colours in Pymol.
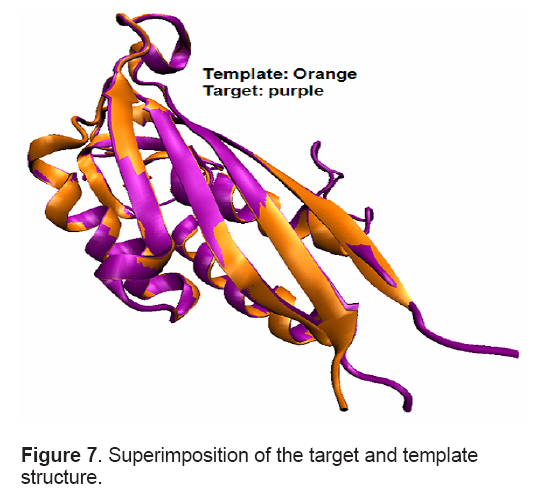
Root-mean-square deviation (RMSD) of 0.05 Å between the backbone atoms of the template and model indicated close homology. This ensures the reliability of the model. Superimposition between target and template structure is shown in Figure 7 .
Q-site finder predicted 10 binding sites in the modeled structure. Out of which the largest site having a volume of 233 cubic Å was selected as active site. Important residues identified in the active site were Val 179,Ile 180,Ala181,Gln 182,Val183,Pro184,Lys185,Ile186,Ser187,Arg190,Val210,Lys 211,Gly212,Lys 213 and Thr 214.
4. Conclusions
Failure to win the long ruthless battle against malaria is mainly attributed to resistance of vectors to insecticides,unhygienic conditions,lack of primary healthcare facilities,limited availability of cheap and effective drugs and above all,the evolution and spread of multi- drug resistant Plasmodium species. This situation has now underscored the need for search of novel drug targets. Often the enzymes critical for the survival of the parasites with no known homolog in the host species are considered as good drug targets as they bring down the risk factors of unwanted side effects elicited by drugs targeting these molecules. MEP pathway houses many good drug targets and among them,MECPS has been projected as one of the most promising and attractive drug target. Structure of this enzyme has been elucidated and explored for the possibility of anti-microbial drug development in other important parasites like Mycobacterium tuberculosis. Effective targeting of this enzyme by possible inhibitors (proved successful in similar pathogens) will pave a way for development of future anti-malarial therapeutics. In the present study,we have constructed a tertiary structure model of MECP synthase of Plasmodium falciparum 3D7. This model will provide an insight about its structure in absence of an experimentally derived crystal structure and will serve as a foundation for rational design of anti-malarial therapeutics.
Acknowledgements
Authors are thankful to the Director of I.I.C.T for his support and encouragement. AKB is thankful to C.S.I.R for Senior Research Fellowship. Authors thank anonymous reviewers for their valuable suggestions for improving the manuscript.
References
- Kohler S.,Delwiche C.F.,Denny P.W.,Tilney L.G.,Webster P.,Wilson R.J.,Palmer J.D.,Roos D.S. (1997) A plastid of probable green algal origin in apicomplexan parasites. Science,275: 1485–1489.
- McFadden G.I.,Waller R.F. (1997) Plastids in parasites of humans. BioEssays,19(11): 1-8.
- Blanchard J.,Hicks J.S. (1999) The nonphotosynthetic plastid in malarial parasites and other apicomplexans is derived from outside the green plastid lineage. J. Euk. Microbiol,46: 367-375.
- Stoebe B.,Kowallik K.V. (1999) Gene-cluster analysis in chloroplast genomics. TIG,15(9): 1-4.
- Roos D.S.,Crawford M.J.,Donald R.G.K.,Kissinger J.C.,Klimczak L.J.,Striepen B. (1999) Origin,targeting,and function of the apicomplexan plastid. Curr. Opin. Microbiol,2: 426-432.
- Fast N.M.,Kissinger J.C.,Roos D.S.,Keeling P.J. (2001) Nuclear-encoded,plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol. Biol. Evol,18: 418-426.
- Wilson R.J.M. (2002) Progress with Parasite Plastids. J.Mol.Biol,319(2): 257-274.
- Gardner M.J. et al. (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature,419: 498–511.
- Steinbacher S.,Kaiser J.,Wungsintaweekul J.,Hecht S.,Eisenreich W.,Gerhardt S.,Bacher A.,Rohdich F. (2002) Structure of 2C-methyl-D -erythritol-2,4- cyclodiphosphate synthase involved in mevalonateindependent biosynthesis of isoprenoids. J.Mol.Biol,316(1): 79-88.
- Roos D.S.,Crawford M.J.,Donald R.G.,Fraunholz M.,Harb O.S.,He C.Y.,Kissinger J.C.,Shaw M.K.,Striepen B. (2002) Mining the Plasmodium genome database to define organellar function: what does the apicoplast do? Philos Trans R Soc Lond B Biol Science,357: 35–46.
- Ralph S.A.,D'Ombrain M.C.,McFadden G.I. (2001) The apicoplast as an antimalarial drug target. Drug Resist Update,4: 145–151.
- McFadden G.I.,Roos D.S. (1999) Apicomplexan plastids as drug targets. Trends in Microbiology,7(8): 328-333.
- Kemp L.E.,Bond C.S.,Hunter W.N. (2002) Structure of 2C-methyl-D-erythritol 2,4 cyclodiphosphate synthase: an essential enzyme for isoprenoid biosynthesis and target for antimicrobial drug development. Proc. Natl Acad. Sci. USA,99: 6591– 6596.
- Bloch K. (1992) Sterol molecule: structure,biosynthesis and function. Steroids,57(8): 378-383.
- Rohmer M.,Knani M.,Simonin P.,Sutter B.,Sahm H. (1993) Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J,295: 517-524.
- Eisenreich W.,Schwarz M.,Cartayrade A.,Arigoni D.,Zenk M.H.,Bacher A. (1998) The deoxyxylulose phosphate pathway of terpenoid biosynthesis in plants and microorganisms. Chem. Biol,5: R221–R233.
- Boucher Y.,Doolittle W.F. (2000) The role of lateral gene transfer in the evolution of isoprenoid biosynthesis pathways. Mol. Microbiol,37: 703–716.
- Couto A.S.,Kimura E.A.,Peres V.J.,Uhrig M.L.,Katzin A.M. (1999) Active isoprenoid pathway in the intra-erythrocytic stages of Plasmodium falciparum: presence of dolichols of 11 and 12 isoprene units. Biochem J,341: 629–637.
- Rohdich F.,Eisenreich W.,Wungsintaweekul J.,Hecht S.,Schuhr C.A.,Bacher A. (2001) Biosynthesis of terpenoids. 2C-Methyl-D-erythritol 2,4- cyclodiphosphate synthase (IspF) from Plasmodium falciparum. Eur. J. Biochem,268: 3190–3197.
- Rodríguez-Concepción M.,Forés O.,Martinez-García J.F.,González V.,Phillips M.A.,Ferrer A.,Boronat A. (2004) Distinct light-mediated pathways regulate the biosynthesis and exchange of isoprenoid precursors during Arabidopsis seedling development. Plant Cell,16(1): 144-156.
- Kuntz L.,Tritsch D.,Grosdemange-Billard C. et al. (2005) Isoprenoid biosynthesis as a target for antibacterial and antiparasitic drugs: phosphonohydroxamic acids as inhibitors of deoxyxylulose phosphate reducto-isomerase. Biochem J,386: 127–135.
- Gasteiger E.,Hoogland C.,Gattiker A.,Duvaud S.,Wilkins M.R.,Appel R.D. et al. (2005) Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook (John M.,Walker),Humana Press.
- Geourjon C.,Deleage G. (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci,11(6): 681–684.
- Cserzö M.,Wallin E.,Simon I.,Von Heijne G.,Elofsson A. (1997) Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng,10(6): 673-676.
- Sonnhammer E.L.L.,Von-Heijne G.,Krogh A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. In Proceedings of the sixth international conference on intelligent systems for molecular biology (Glasgow T.L.J.,Major F.,Lathrop R.,Sankoff D.,Sensen C.),AAAI Press,Menlo Park,CA,175–182.
- Hirokawa T.,Boon-Chieng S.,Mitaku S. (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics,14(4): 378–379.
- Hofmann K.,Stoffel W. (1993) TMbase - A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler,374: 166.
- Tusnády G.E.,Simon I. (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics,17(9): 849-850.
- Berman H.M.,Westbrook J.,Feng Z.,Gilliland G.,Bhat T.N.,Weissig H.,Shindyalov I.N.,Bourne P.E. (2000) The Protein Data Bank. Nucleic Acids Res,28(1): 235–242.
- Thompson J.D.,Gibson T.J.,Plewniak F.,Jeanmougin F.,Higgins D.G. (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res,24: 4876–4882.
- Sali A.,Blundell T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol,234(3): 779–815.
- DeLano W.L. (2002) The PyMOL Molecular Graphics System on World Wide Web. (https://www.pymol.org).
- Humphrey W.,Dalke A.,Schulten K. (1996) VMD - Visual Molecular Dynamics. J. Mol. Graph Mod,14: 33-38.[26] Hirokawa T.,Boon-Chieng S.,Mitaku S. (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics,14(4): 378–379.
- Hofmann K.,Stoffel W. (1993) TMbase - A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler,374: 166.
- Tusnády G.E.,Simon I. (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics,17(9): 849-850.
- Berman H.M.,Westbrook J.,Feng Z.,Gilliland G.,Bhat T.N.,Weissig H.,Shindyalov I.N.,Bourne P.E. (2000) The Protein Data Bank. Nucleic Acids Res,28(1): 235–242.
- Thompson J.D.,Gibson T.J.,Plewniak F.,Jeanmougin F.,Higgins D.G. (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res,24: 4876–4882.
- Sali A.,Blundell T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol,234(3): 779–815.
- DeLano W.L. (2002) The PyMOL Molecular Graphics System on World Wide Web. (https://www.pymol.org).
- Humphrey W.,Dalke A.,Schulten K. (1996) VMD - Visual Molecular Dynamics. J. Mol. Graph Mod,14: 33-38.
- Lovell S.C.,Davis I.W.,Arendall III W.B.,De Bakker P.I.W.,Word J.M.,Prisant M.G.,Richardson J.S.,Richardson D.C. (2003) Structure Validation by Cα Geometry: φ,ψ and Cβ Deviation. Proteins: Structure,Function and Genetics,50: 437-450.
- Laurie A.T.,Jackson R.M. (2005) Q-Site Finder: an energy-based method for the prediction of proteinligand binding sites. Bioinformatics,21: 1908-1916.
- Mamatha D.M.,Nagalakshmamma K.,Rajesh D.V.A.,Sheerin V.S. (2007) Protein modeling of apical membrane antigen- 1(AMA-1) of Plasmodium cynomolgi. Afr. J. Biotechnol,6(22): 2628–2632.
- Kumar R.,Pavithra S.R.,Tatu U. (2007) Threedimensional structure of heat shock protein 90 from Plasmodium falciparum: molecular modelling approach to rational drug design against malaria. J Biosci,32(3): 531–536.
- Rastelli G.,Pacchioni S.,Sirawaraporn W.,Sirawaraporn R.,Parenti M.D.,Ferrari A.M. (2003) Docking and Database Screening Reveal New Classes of Plasmodium falciparum Dihydrofolate Reductase Inhibitors. J. Med. Chem,46(14): 2834– 2845.
- Banerjee A.K.,Arora N.,Murty U.S.N. (2009) Structural model of the Plasmodium falciparum Thioredoxin reductase: A novel target for antimalarial drugs. Journal of vector borne diseases,46(3): 171- 183.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences