Thymoquinone Regulates Gene Expression Levels in Morphine Addiction Pathways in Opioid Receptor Expressing Cells (U87 MG)
Adnan LH, Mohamad N, Mat KC, Yeo CC, Bakar NHA, Ismail R
1Faculty of Medicine, Universiti Sultan Zainal Abidin (UniSZA), Terengganu, Malaysia
2Opioid Research Interest Group, Faculty of Medicine, Universiti Sultan Zainal Abidin (UniSZA), 20400, Kuala Terengganu, Terengganu, Malaysia.
Received date: April 22, 2017; Accepted date: May 10, 2017; Published date: May 17, 2017
Citation: Adnan LHM, Mohamad N, Mat KC, et al. Thymoquinone Regulates Gene Expression Levels in Morphine Addiction Pathways in Opioid Receptor Expressing Cells (U87 MG). Electronic J Biol, 13:2
Abstract
Background: New drugs are continuously being developed for the treatment of opioid dependence individuals. Thymoquinone is one of the drugs that exhibits inhibitory characteristics based on in vivo and in vitro models. Aim: This study further investigates the effects of thymoquinone on human global gene expression from opioid receptor expressing cell line (OREC) using RNA sequencing technology. The cell line that we used was human monoglioma cancer cells (U87 MG). Method: The quantification of RNA samples was carried out using an Agilent 2100 Bioanalyser to determine the RNA integrity number (RIN). Samples with RIN>9.4 were used for further analysis. The universal human reference RNA was used as the common reference. The samples were treated with morphine alone (35 μM), morphine with methadone (162 μM), morphine with methadone and TQ (61 μM) and morphine with TQ for 48 h. The control cells were treated with an equivalent volume of vehicle in growth media. Total RNA was isolated from the U87 MG cells using the RNA mini kit’s protocol (Geneaid) and quantified using a BioDrop 2000c spectrophotometer. After mRNA enrichment and cDNA synthesis, the amplified cDNA fragments were paired-end sequencing using Illumina sequencing system with 2*150 sequencing method and subjected to subsequent bioinformatics analysis. Findings: The results showed that thymoquinone upregulated phosphodiesterase 1 A (PDE1A) genes, gamma-aminobutyric acid type A receptor theta subunit (GABRQ) and G protein subunit beta 3 (GNG3) genes in which these genes were down regulated by the chronic morphine.
Keywords
Thymoquinone; U87MG; RNAsequencing; Morphine addiction.
1. Introduction
The World Drug Report 2016 had reported that an estimate 247 million people used drugs and 29 million people suffered from drug use disorders but only one in 6 people with drug use disorders is in treatment [1]. In Malaysia, recent Statistics from National Anti-Drug Agency showed that in December 2015, as many as 26, 668 drug users were detected in 2015. Most of the type of drugs that are currently being abused is opioid-type of drugs with more than 60% of the drug addicts used Heroine and Morphine compared to other types of drugs [2].
Thymoquinone (TQ) is an opioid receptor stimulating compound reported to have medicinal potentials in numbers of illnesses including the treatment of opioid dependence [3]. The inhibitory effects of thymoquinone on opioid dependence and withdrawal syndromes have been reported in human subjects, animal studies and recently at cellular levels derived from monoglioma cancer cells U87 MG. U87 MG has been reported to actively express opioid receptors [4]. Many researches regarding opioid dependence had been conducted using this Opioid Receptor Expressing cells (OREC) or U87 MG cells in order to unreveal the exact mechanism of several candidates of drugs [4,5].
However, to date, it is our first attempt to investigate the effects of thymoquinone in opioid dependence using RNA Sequencing technologies. The inhibitory activity of thymoquinone has been reported to attenuate the development of opioid tolerance and dependence by inhibiting brain oxidative stress, inducible nitric oxide synthase expression and attenuating morphine-induced cyclic Adenosine Monophosphate (cAMP) overshoot in mice and OREC cells [5,6]. These findings were based on animal behavioural study, biochemical tests, real time polymerase chain reaction (RT-PCR) and Enzyme- Linked Immunosorbent Assay (ELISA). Therefore, with the advent of Next Generation Sequencing (NGS) technology, further investigation is required to determine its effects on human genome expression using RNA Sequencing (RNA-seq) technique.
2. Materials and Methods
2.1 Complete cell culture medium preparation
RPMI 1640 (Invitrogen, Gibco, Carlsbad, CA, USA) medium, fetal bovine serum (FBS) and penicillin/ streptomycin were used to culture the OREC cell line U87 MG (ATCC® HTB-14™).
2.2 Thymoquinone solution preparation
Thymoquinone (Sigma-Aldrich, Saint-Quentin- Fallavier) was dissolved in 0.1% DMSO (Merck, Germany) to make up stock concentration of 305 mm (50 mg/mL). All samples was prepared and diluted freshly prior to experiment.
2.3 Cell culture and treatment
The U87 MG cells were seeded at 5 × 106 cells/ flask in tissue culture flasks. They were cultured at 0.5% CO2 in a humidified incubator at 37°C (Thermo Scientific, Waltham, MA, USA) until it reaches 60- 70% confluency. The control cells were treated with an equivalent volume of vehicle in growth media. Four biological replicates from each sample were prepared in separate culture flasks. The samples were treated with morphine alone (35 μM), morphine with methadone (162 μM), morphine with methadone and TQ (61 μM) and morphine with TQ for 48 h. The Phosphate Buffer Saline (PBS) and Trypsin (0.10- 0.25%) solution were used to detach the cells from the flask surface. The cells were then centrifuged at 300x g for 10 min. The supernatant was removed and the cells were washed with RBC Lysis Buffer by pipette following the total RNA mini kit’s protocol (Geneaid).
2.4 RNA isolation
Total RNA was isolated from the U87 MG cells using the RNA mini kit’s protocol (Geneaid) according to the manufacturer's instructions. The quantity of RNA was measured using a spectrophotometer (BioDrop 2000c; Thermo Scientific). Samples with the RNA concentration (A260/A280 ≥ 1.8 ng/μl) and purity (A230/A260 ≥ 2.0 ng/μl) were selected. An Agilent 2100 Bioanalyser was used to determine the RNA integrity number (RIN). The degradation level was identified using the RNA 6000 Nano LabChip kit (Agilent Technologies, Santa Clara, CA, USA). The samples with RIN>9.4 were selected for further analysis.
2.5 mRNA enrichment by Oligo(dT)
Oligo(dT) was paired with the ployA tail in eukaryotic mRNA 3' region and was used to isolate mRNA from the total RNA for the analysis of the transcriptome. Using metal ions, mRNAs were fragmented into short fragments (about 200 bp) randomly.
2.6 cDNA synthesis and adaptor ligation
mRNA fragments were transcribed into first-strand cDNA using reverse transcriptase and random primers, followed by second strand cDNA synthesis. The double-stranded cDNA was further performed with end repaired, A base tailed and indexed adapters were ligated.
2.7 Illumina sequencing
cDNA library was amplified using DNA polymerase for 15 PCR cycles. Library were recovered by gel extraction for target fragments selected on 2% Low Range Ultra Agarose (Certified Low Range Ultra Agarose) and later Quantified by TBS380 (Picogreen). PCR amplification were bridged using the cBot Cluster Generation System and later the amplified cDNA fragments were paired-end sequencing using Illumina sequencing system with 2*150 sequencing method.
2.8 Bioinformatics analysis reports
After Illumina PE library (300 bp) was constructed, Illumina platform was adopted to complete the 2 × 150 bp sequencing. Primary sequencing data (raw reads) then was subjected to Quality Control (QC) to determine if a sequencing read is qualified or not. At last, raw reads were filtered into clean reads and subjected to subsequent bioinformatics analysis.
3. Results and Discussion
3.1 Quality shear and statistics of sequencing data
The original sequencing data will contain primers, low quality reads or the reads with high N rate or too short reads, which seriously affect the quality of subsequent analysis. In order to ensure the accuracy of subsequent biological information analysis, we conduct the raw sequencing data quality control (QC), so as to obtain the sequencing data with high quality (the clean data) to ensure the smooth progress of the follow-up analysis (Table 1). Results from the clean data statistics showed that each of the samples has good base call accuracy. Q scores of 30 (Q30%) is considered as a benchmark for quality in next-generation sequencing. When the sequencing quality reaches Q30, virtually all of the reads from our samples will be perfect having zero errors and ambiguities. This means that the base call accuracy is 99.9% (the probability of a correct base call). A lower base call accuracy of 99% (Q20%) will have an incorrect base call probability of 1 in 100, meaning that every 100bp sequencing reads will likely contain an error. The percentage of GC contents in our samples also at optimum range with more than 50%.
3.2 Mapping reads to reference genome
We used TopHat as a tool for mapping reads. TopHat is a fast splice junction mapper for RNA-Seq reads. It aligns RNA-Seq reads to mammalian-sized genomes using the ultra-high throughput short read aligner Bowtie, and then analyzes the mapping results to identify splice junctions between exons. Tophat2 includes three main steps: transcriptome alignment (optional), genome alignment and spliced alignment. In general, we use the default parameters, and we will also change the parameters if necessary according to the organism gene model. Our results showed that all of our samples (>89%) were totally mapped to the human genome as our reference genome (Table 2).
| Sample Name/Clean Data | Sequence Number | Base Number (bp) | Error% | Q20% | Q30% | GC% |
|---|---|---|---|---|---|---|
| MM | 23761402 | 3457818145 | 0.0163 | 96.82 | 90.93 | 50.39 |
| MMT MT |
24120562 26572346 |
3506764009 3871829885 |
0.0165 0.0162 |
96.75 96.87 |
90.75 91.04 |
51.15 51.09 |
| MOR | 24914480 | 3628597378 | 0.0164 | 96.79 | 90.84 | 51.25 |
| UT | 37847876 | 5493447539 | 0.0118 | 97.69 | 93.74 | 50.29 |
Q20, Q30%: Percentage of bases which phred value greater than 20, 30
Error%: The error rate of bases
GC%: Percentage of both G and C bases
UT: Untreated Cells; MOR: Morphine-Treated Cells; MM: Morphine and Methadone; MMT: Morphine, Methadone and Thymoquinone-Treated Cells; MT: Morphine and Thymoquinone-Treated Cells
Table 1. Clean data statistics.
| Sample Name | UT | MOR | MM | MT | MMT |
|---|---|---|---|---|---|
| Total Reads | 37847876 | 24914480 | 23761402 | 26572346 | 24120562 |
| Total mapped | 33927033 (89.64%) | 22185531 (89.05%) | 21983133 (92.52%) | 24557200 (92.42%) | 2251026 (92.25%) |
| Multiple Mapped | 433992 (1.15%) | 292276 (1.17%) | 309734 (1.30%) | 342958 (1.29%) | 322826 (1.34%) |
| Uniquely Mapped | 33493041 (88.49%) | 21893255 (87.87%) | 21673399 (91.21%) | 24214242 (91.13%) | 21928200 (90.91%) |
| Left Mapped | 18022141 (47.62%) | 11401362 (45.76%) | 11300111 (47.56%) | 12617984 (47.49%) | 11457492 (47.50%) |
| Right Mapped | 15904892 (42.02%) | 10784169 (43.28%) | 10683022 (44.96%) | 11939216 (44.93%) | 10793534 (44.75%) |
UT: Untreated Cells; MOR: Morphine-Treated Cells; MM: Morphine and Methadone; MMT: Morphine, Methadone and Thymoquinone-Treated Cells; MT: Morphine and Thymoquinone-Treated Cells.
Table 2. Mapping results.
| Gene Symbol | Description | log2FC | p-value | FDR |
|---|---|---|---|---|
| ADCY7 | Adenylate cyclase 7 | 1.65 | 2.16E-12 | 1.58E-11 |
| PDE4D | Phosphodiesterase 4D | 1.34 | 4.23E-14 | 3.57E-13 |
log2FC: The fold change difference compared to control (untreated) cells in logarithm base 2
FDR: Corrected p-value
Significant: Significant or not in the criteria (FDR<=0.05|log2FC|>=1).
Table 3. List of genes which were significantly upregulated following chronic treatment with morphine
3.3 Differential expression analysis
EdgeR or deseq2 is used for differential expression analysis. Gene read count data is calculated as the input of EdgeR or deseq2. This analysis method is based on the negative binomial distribution model. The significantly differentially expressed genes screening criteria is: FDR<0.05 && | log2FC | >=1.
Comparison of chronic morphine with control (untreated) conditions revealed significant upregulation of Adenylate cyclase 7 (ADCY7) and Phosphodiesterase 4D genes (PDE4D) (Table 3) and at the same situation, chronic morphine treatment for 48 hours significantly downregulated 10 transcripts as listed in the Table 4. Most of the downregulated genes are the genes that are mainly involved in cAMP signalling pathway and are important in inhibitory of neurotransmitter release such as dopamine (DA).
Meanwhile, Methadone receiving samples exhibited significantly upregulation of 5 transcripts (Table 5) and downregulation of 7 transcripts in morphine addiction pathway (Table 6). Chronic morphine treatment had caused the downregulation of phosphodiesterase 1A (PDE1A) and G protein subunit beta 3 (GNB3) genes. However, co-treatment of the cells with methadone had significantly upregulated both of the genes previously downregulated by morphine (Table 5).
When the cells were co-treated with thymoquinone, there were 5 transcripts that were significantly upregulated and 7 transcripts were significantly downregulated as compared to morphine-treated cells alone (Tables 7 and 8). In this sample, our results showed that thymoquinone also significantly upregulated PDE1A and GNB3 genes like methadone. Interestingly, it also induce the upregulation of Gamma-aminobutyric acid type A receptor theta subunit (GABRQ) instead of PDE1A and GNB3 genes, where these genes had been downregulated following chronic morphine treatment (Table 8).
| Gene Symbol | Description | log2FC | p-value | FDR |
|---|---|---|---|---|
| PDE1A | Phosphodiesterase 1A | -3.32 | 1.23E-09 | 6.93E-09 |
| CACNA1A | Calcium voltage-gated channel subunit alpha1 A | -1.16 | 5.34E-30 | 1.10E-28 |
| GNB4 | G protein subunit beta 4 | -1.04 | 7.14E-18 | 8.01E-17 |
| PDE7B | Phosphodiesterase 7B | -1.74 | 3.17E-03 | 7.17E-03 |
| PDE3A | Phosphodiesterase 3A | -5.33 | 1.57E-13 | 1.27E-12 |
| GABRQ | Gamma-aminobutyric acid type A receptor theta subunit | -1.89 | 2.85E-26 | 5.02E-25 |
| GNB3 | G protein subunit beta 3 | -1.22 | 8.00E-04 | 2.04E-03 |
| CDCA3 | Cell division cycle associated 3 | -2.67 | 1.02E-55 | 4.61E-54 |
| GNG2 | G protein subunit gamma 2 | -1.23 | 7.37E-14 | 6.11E-13 |
| GABRA3 | Gamma-aminobutyric acid type A receptor alpha3 subunit | -2.3 | 2.91E-39 | 8.61E-38 |
log2FC: The fold change difference compared to control (untreated) cells in logarithm base 2 FDR: Corrected p-value
Significant: Significant or not in the criteria (FDR<=0.05|log2FC|>=1).
Table 4. List of genes which were significantly downregulated following chronic treatment with morphine.
| Gene Symbol | Description | log2FC | p-value | FDR |
|---|---|---|---|---|
| PDE1A | Phosphodiesterase 1A | 3.17 | 1.30E-08 | 6.93E-08 |
| GABBR1 | Gamma-aminobutyric acid type B receptor subunit 1 | 1.15 | 3.25E-12 | 2.58E-11 |
| PRKACB | Protein kinase cAMP-activated catalytic subunit beta | 1.17 | 1.04E-29 | 2.79E-28 |
| GNB3 | G protein subunit beta 3 | 1.13 | 2.18E-03 | 5.23E-03 |
| GNG7 | G protein subunit gamma 7 | 1.78 | 1.52E-02 | 2.99E-02 |
log2FC: The fold change difference compared to morphine-treated cells in logarithm base 2
FDR: Corrected p-value
Significant: Significant or not in the criteria (FDR<=0.05|log2FC|>=1).
Table 5. List of genes which were significantly upregulated following treatment with methadone.
| Gene Symbol | Description | log2FC | p-value | FDR |
|---|---|---|---|---|
| GNAI1 | G protein subunit alpha i1 | -1.08 | 2.52E-17 | 3.07E-16 |
| PDE4B | Phosphodiesterase 4B | -2.13 | 1.86E-11 | 1.36E-10 |
| GNG11 | G protein subunit gamma 11 | -1.12 | 3.99E-41 | 2.02E-39 |
| CDCA3 | Cell division cycle associated 3 | -1.38 | 1.12E-05 | 4.04E-05 |
| GNG2 | G protein subunit gamma 2 | -2.44 | 6.47E-17 | 7.53E-16 |
| ADORA1 | Adenosine A1 receptor | -2.01 | 4.94E-18 | 6.36E-17 |
| GABRA3 | Gamma-aminobutyric acid type A receptor alpha3 subunit | -2.03 | 4.14E-09 | 2.35E-08 |
log2FC: The fold change difference compared to morphine-treated cells in logarithm base 2
FDR: Corrected p-value
Significant: Significant or not in the criteria (FDR<=0.05|log2FC|>=1).
Table 6. List of genes which were downregulated following treatment with methadone.
| Gene Symbol | Description | log2FC | p-value | FDR |
|---|---|---|---|---|
| ARRB1 | Arrestin beta 1 | -1.08 | 1.16E-05 | 4.92E-05 |
| PDE4B | Phosphodiesterase 4B | -1.46 | 3.23E-07 | 1.76E-06 |
| CDCA3 | Cell division cycle associated 3 | -1.36 | 2.00E-05 | 8.12E-05 |
| PDE1C | Phosphodiesterase 1C | -1.98 | 4.55E-03 | 1.12E-02 |
| GNG2 | G protein subunit gamma 2 | -1.99 | 1.66E-13 | 1.90E-12 |
| ADORA1 | Adenosine A1 receptor | -2.73 | 2.76E-25 | 7.78E-24 |
| GABRA3 | Gamma-aminobutyric acid type A receptor alpha3 subunit | -1.48 | 3.72E-06 | 1.71E-05 |
log2FC: The fold change difference compared to morphine-treated cells in logarithm base 2
FDR: Corrected p-value
Significant: Significant or not in the criteria (FDR<=0.05|log2FC|>=1).
Table 7. List of genes which were significantly downregulated following treatment with thymoquinone
| Gene Symbol | Description | log2FC | p-value | FDR |
|---|---|---|---|---|
| PDE1A | Phosphodiesterase 1A | 2.43 | 1.15E-04 | 4.06E-04 |
| GABBR1 | Gamma-aminobutyric acid type B receptor subunit 1 | 1.05 | 4.04E-10 | 3.28E-09 |
| GNG7 | G protein subunit gamma 7 | 2.18 | 5.89E-04 | 1.80E-03 |
| GABRQ | Gamma-aminobutyric acid type A receptor theta subunit | 1.25 | 6.00E-11 | 1.39E-09 |
| GNB3 | G protein subunit beta 3 | 1.07 | 4.13E-03 | 1.55E-02 |
log2FC: The fold change difference compared to morphine-treated cells in logarithm base 2
FDR: Corrected p-value
Significant: Significant or not in the criteria (FDR<=0.05|log2FC|>=1).
Table 8. List of genes which were significantly upregulated following treatment with thymoquinone.
3.4 DEGs cluster analysis
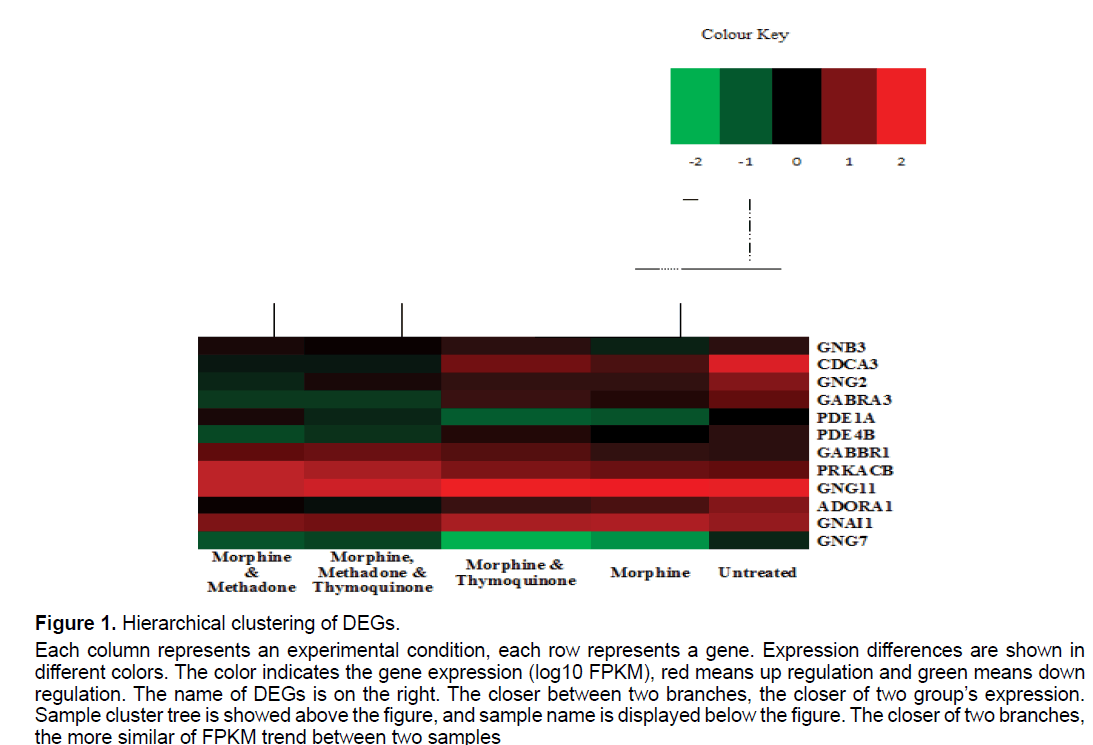
Genes with similar expression patterns usually have similar function and grouped together within a cluster which are co-regulated and functionally related. We perform cluster analysis of gene expression patterns by distance calculation algorithm using spearman among the samples. Clustering method used is hierarchical methods. Here we noticed that morphinetreated samples and co-treatment of morphine with thymoquinone had a similar expression profile, which linked to form a group. A similar pattern was observed in the co-treatment of morphine with methadone (MM) and methadone with thymoquinone-treated (MMT) samples (Figure 1). The heatmap showed are based on the log10 FPKM values for each of the individual samples in this study. Based on the DEGs cluster analysis, G protein subunit gamma 11 (GNG11), gamma-aminobutyric acid type B receptor subunit 1 (GABBR1), protein kinase cAMP-activated catalytic subunit beta (PRKACB) and G protein subunit alpha i1 (GNAI1) are among the most similar genes that gave similar expression patterns and functions across all of the samples (Figure 1).
4. Discussion
Results of the present study showed that chronic morphine incubations in OREC (U87 MG) cell lines had led to generate specific and more global differential expression of the genes. Each of the drug treatment group except for untreated group was compared directly with morphine-treated samples, which were handled and treated at the same time, under the same conditions. Total RNA were isolated and labelled, and the sequences were run at the same time for each experiment. Among the genes regulated in the OREC by chronic morphine administrations are essential for cAMP signalling pathway, and also involved in neurotransmission release such as Dopamine (DA) release including Phosphodiesterase (PDE), G protein subunit (G i/o) and Gamma-Aminobutyric Acid (GABA).
Figure 1:Hierarchical clustering of DEGs.
Each column represents an experimental condition, each row represents a gene. Expression differences are shown in
different colors. The color indicates the gene expression (log10 FPKM), red means up regulation and green means down
regulation. The name of DEGs is on the right. The closer between two branches, the closer of two group’s expression.
Sample cluster tree is showed above the figure, and sample name is displayed below the figure. The closer of two branches,
the more similar of FPKM trend between two samples
Morphine was significantly upregulated Adenylate cyclase 7 (ADCY 7) and Phosphodiesterase 4D (PDE4D) genes when compared with the untreated samples. This ADCY7 gene encodes a membranebound adenylate cyclase that catalyses the formation of cyclic AMP from ATP and is inhibitable by calcium. A particular emphasis has been given to the role of cAMP during opioid dependency [7]. An upregulation of this ADCY 7 gene had been shown to correlate with the chronic actions of the morphine in our morphine treated samples [8]. An upregulation of this gene also had been observed in another ex-vivo study of brain region-specific gene expression in mice exposed with the chronic stressors, suggesting the association of this gene with genetic depression caused by the chronic morphine [9]. Phosphodiesterase 4D gene is responsible for the degradation of cAMP through the activity of phosphodiesterases that can be found throughout the brain [10]. Morphine withdrawal upregulates PDE4D gene expression level in the absence of PDE4D inhibitors [11]. Thus, our results are in consistent with the findings from the literatures in which the upregulation of this gene in morphine-treated sample was known to be caused by morphine induced withdrawal as opposed to the untreated sample.
Co-treatment of morphine with methadone revealed more significantly up regulated genes such as phosphodiesterase 1A (PDE1A), gammaaminobutyric acid type B receptor subunit 1 (GABBR1), protein kinase cAMP-activated catalytic subunit beta (PRKACB), G protein subunit beta 3 (GNB3) and G protein subunit gamma 7 (GNG7) genes. PDE1A is Ca(2+)/calmodulin-dependent phosphodiesterases (PDEs) (CaM-PDEs) that are activated by calmodulin in the presence of Ca(2+) [12,13]. Human PDE1A genes are responsible for modulating cellular levels of cAMP and cGMP in response to stimuli, considering the importance of cAMP and cGMP to disparate physiological functions including opioid withdrawal [14]. GABBR1 gene encodes a receptor for gamma-aminobutyric acid (GABA), which is the main inhibitory neurotransmitter in the mammalian central nervous system. To date, an association between the regulations of this gene with opioid dependence is still remain unclear except for alcoholics, where the GABBR1 splicing is altered [15]. Another recent study had shown that the complex formation of the GABBR1 subunit and G protein-coupled receptor kinase (GRK) 4 or 5 can contribute to the desensitization of GABBR1 in mechanism of morphine tolerance [16].
Protein kinase cAMP-activated catalytic subunit beta (PRKACB) gene is a gene that encodes for a protein that acts as a catalytic subunit of cAMP (cyclic AMP)- dependent protein kinase, which mediates signalling though cAMP. cAMP signaling is important to a number of processes, including cell proliferation and differentiation. PRKACB can induce phosphorylation of cAMP response element binding protein (CREB), a transcription factor that may regulate neuroadaptations related to morphine dependence [17]. Upregulation of PRKACB in this sample would increase the phosphorylation of CREB, which had been demonstrated to happen during morphine withdrawal after chronic morphine treatment [18].
Heterotrimeric G proteins are composed of three subunits, which are an alpha, beta and gamma. G proteins act as an intermediary path in signal transductions from the receptors involved in substance dependence and effector proteins such as adenylyl cyclase isoforms, phospholipase isoforms, ion channels, protein tyrosine kinases, and mitogenactivated protein kinases (MAPKs) [19]. A singlenucleotide polymorphism (C825T) existed in this gene and GNB 3 C825T polymorphism is associated with several disorders including hypertension, obesity, and major depressive disorder. An association between the polymorphism of this gene had been shown to affect major depressive disorder in alcohol dependence study, supporting the hypothesis of the involvement of G-proteins in mood regulation [20]. An upregulation of GNB 3 gene would be expected to increase the regulation of mood depression associated with morphine dependence.
In other samples, when TQ is co-treated with Methadone and Morphine, it results in a significantly upregulation of PDE gene, GABA and G protein subunit. PDEs are classified into classes I, II and III [21]. In our results, Phosphodiesterase 1A gene (PDE1A) also had been significantly upregulated by TQ instead of methadone as compared to morphine alone. This gene encodes for Ca2+/CaMdependent cAMP- and cGMP-hydrolyzing PDEs. In humans, PDE1A shows high affinity for cGMP [21]. In morphine withdrawal mechanism where an increased in adenosine tone is taking place, upregulation of PDE genes eventually led to the upregulation of Adora 1 gene (A1) which in turn induce endocytosis of Dopamine (DA) by the upregulations of GABA genes and G protein subunit. Thus, this will cause the disinhibition of dopamine cell firing that will enhance the release of dopamine from the terminals. Apart from these cellular effects, it will contribute to the persistent of behavioural change among opioid dependence individuals [21].
Interestingly, when we co-treated TQ alone with Morphine, it revealed only two significantly upregulated genes which are GABA genes and G protein subunits. The GABA genes subunit involved is gamma-aminobutyric acid type A receptor theta subunit or GABRQ and G protein subunit beta 3 (GNB3). However, the changes in the regulation of these genes are not enriched in morphine addiction pathway like in other samples. This is due to the GABRQ gene encodes the theta subunit of the GABA A receptor and also believed to be the candidate genes of two different neurologic diseases: earlyonset parkinsonism (Waisman syndrome) and X-linked mental retardation (MRX3) [Provided by RefSeq, Nov 2009], whereas, the GNB3 gene encodes for beta subunit of G proteins that are important as regulators for alpha subunits, as well as of certain signal transduction receptors and effectors. A single-nucleotide polymorphism (C825T) in this gene is associated with essential hypertension, obesity and the occurrence of the splice variant GNB3-s, which appears to have increased activity. GNB3-s is an example of alternative splicing caused by a nucleotide change outside of the splice donor and acceptor sites [provided by RefSeq, Jul 2014].
5. Conclusion
As a conclusion, our findings suggest that thymoquinone acts in a synergistic manner with methadone involving the morphine addiction pathways as indicated by our RNA-sequencing results. The co-administration of thymoquinone and methadone still requires further investigation in order to fully elucidate the treatment outcomes in methadone maintenance therapy (MMT). It is noted that RNA-sequencing results are accepted measures of gene expression levels and not only that, it can measure low-expressed genes, alternative splice variants, and novel transcripts that makes RNASequencing a far more precise measurement of levels of transcripts and their isoforms than other methods [22-24]. RNA-sequencing experiments also produce robust differential analysis in at transcript resolution, revealing a layer of regulation not readily observable with other high-throughput technologies [24]. Nevertheless, the results obtained in this study could perhaps be further validated by quantitative real-time reverse transcriptase PCR.
6. Acknowledgement
This study was funded by Niche Research Grant Scheme (NRGS) from the Ministry of Education (MOE), Malaysia RR057-1 Universiti Sultan Zainal Abidin. The authors would like to extend their appreciation to the statistician for bioinformatic analysis.
References
- World Drug Report. (2016). United Nations Office on Drugs and Crime, New York. United Nation. ISBN: 978-92-1-148286-7.
- Drug information. (2015). National Anti-drug Agency, Ministry of internal affairs, Malaysia.
- Nutten S, Philippe D, Mercenier A, et al. (2012). Opioid receptors stimulating compounds (thymoquinone, Nigella sativa) and food allergy: Google Patents.
- Byrne LS, Peng J, Sarkar S,et.al. (2012). Interleukin-1 beta-induced up-regulation of opioid receptors in the untreated and morphine-desensitized U87 MG human astrocytoma cells. J Neuroinfl. 9: 252.
- Adnan LHMA, Mohamad N, Mat KC, et.al. (2016). The Effect of Thymoquinone on concentration of human mu-opioid receptors mediated by chronic morphine treatment in opioid receptor expressing cell (U87 MG). Acta Bioethica. 22.
- Abdel-Zaher AO, Mostafa MG, Farghly HM, et.al. (2013). Inhibition of brain oxidative stress and inducible nitric oxide synthase expression by thymoquinone attenuates the development of morphine tolerance and dependence in mice. Eur J Pharmacol. 702: 62-70.
- Hanoune J, Defer N. (2001). Regulation and role of adenylyl cyclase isoforms. Annu Rev Pharmacol Toxicol. 41: 145-174.
- Yoshimura M, Wu PH, Hoffman PL, et.al. (2000). Overexpression of type 7 adenylyl cyclase in the mouse brain enhances acute and chronic actions of morphine. Mol Pharmacol. 58: 1011-1016.
- Postmortem G. (2014). Current status of the human evidence for the genetics of depression. Behavioral Genetics of the Mouse. Genetic Mouse Models of Neurobehavioral Disorders. 2: 254.
- Thompson BE, Sachs BD, Kantak KM, et.al. (2004). The Type IV phosphodiesterase inhibitor rolipram interferes with drug‐induced conditioned place preference but not immediate early gene induction in mice. Eur J Neurol. 19: 2561-2568.
- Hamdy MM, Mamiya T, Noda Y, et al. (2001). A selective phosphodiesterase IV inhibitor, rolipram blocks both withdrawal behavioral manifestations and c-Fos protein expression in morphine dependent mice. Behav Brain Res. 118: 85-93.
- Michibata H, Yanaka N, Kanoh Y, et.al. (2001). Human Ca2+/calmodulin-dependent phosphodiesterase PDE1A: Novel splice variants, their specific expression, genomic organization, and chromosomal localization. Biochim Biophys Acta. 1517: 278-287.
- Fidock M, Miller M, Lanfear J. (2002). Isolation and differential tissue distribution of two human cDNAs encoding PDE1 splice variants. Cell Signal. 14: 53-60.
- Bingham J, Sudarsanam S, Srinivasan S. (2006). Profiling human phosphodiesterase genes and splice isoforms. Biochem Biophys Res Commun. 350: 25-32.
- Lee C, Mayfield RD, Harris RA. (2014). Altered gamma-aminobutyric acid type B receptor subunit 1 splicing in alcoholics.Biol Psychiatry. 75: 765-773.
- Ando Y, Hojo M, Kanaide M, et.al. (2011). S(+)-ketamine suppresses desensitization of gamma-aminobutyric acid type B receptor-mediated signaling by inhibition of the interaction of gamma-aminobutyric acid type B receptors with G protein-coupled receptor kinase 4 or 5. Anesthesiology. 114: 401-411.
- Rashid F, Shah A, Shan G. (2016). Long non-coding RNAs in the cytoplasm. Geno Proteo Bioinf. 14: 73-80.
- Chartoff EH, Papadopoulou M, Konradi C, et.al. (2003). Dopamine-dependent increases in phosphorylation of cAMP response element binding protein (CREB) during precipitated morphine withdrawal in primary cultures of rat striatum. J Neurochem. 87: 107-118.
- Khan SM, Sleno R, Gora S, et.al. (2013). The expanding roles of Gβγ subunits in G protein–coupled receptor signaling and drug action. Pharmacol Rev. 65: 545-577.
- Prestes A, Marques F, Hutz M, et.al. (2007). The GNB3 C825T polymorphism and depression among subjects with alcohol dependence. J Neural Transm. 114: 469-472.
- Omori K, Kotera J. (2007). Overview of PDEs and their regulation. Circ Res. 100: 309-327.
- Marioni JC, Mason CE, Mane SM, et.al. (2008). RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18:1509-1517.
- Wang Z, Gerstein M, Snyder M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 10: 57-63.
- Trapnell C, Hendrickson DG, Sauvageau M, et.al. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 31: 46-53.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences