Three New/Old Vertex-Degree-Based Topological Indices of Some Dendrimers Structure
Adnan Aslam, Yasir Bashir, Muhammad Rafiq, Faiza Haider, Nazeer Muhammad, Nargis Bibi
Adnan Aslam1, Yasir Bashir2, Muhammad Rafiq2, Faiza Haider2, Nazeer Muhammad2,4,*, Nargis Bibi3
1Department of Natural Sciences and Humainities, University of Engineering and Technology, Lahore (RCET), Pakistan
2Department of Mathematics, COMSATS Institute of Information Technology, Wah Cantt, Pakistan
3Department of Computer Science, Fatima Jinnah Women University, Pakistan
4Department of Applied Mathematics, Hanyang University, South Korea
- *Corresponding Author:
- Tel: 00923320568444
Email: nazeer@hanyang.ac.kr
Received date: February 21, 2017; Accepted date: March 23, 2017; Published date: March 30, 2017
Citation: Aslam A, Bashir Y, Rafiq M, et al. Three New/Old Vertex-Degree-Based Topological Indices of Some Dendrimers Structure. Electronic J Biol, 13:1
Abstract
There is a natural linkage between the molecular structures and the bio-medical and pharmacology characteristics. A topological index can be considered as transformation of chemical structure in to real number and has been used as a predictor parameter. There are certain vertex-degree-based topological indices which has been used extensively in the chemical graph theory but recently no further attention is given to these topological indices. These are the reciprocal Randi c′ index (RR), the reduced reciprocal Randi c’ index (RRR) and the reduced second Zagreb index RM2. In this paper we determine reciprocal Randi c’ index (RR), the reduced second Zagreb index RM2, and the reduced reciprocal Randic’ index (RRR) of Poly (Propyl) Ether Imine, porphyrin and Zinc-Porphyrin dendrimers.
Keywords
Randi c’ index; Zagreb index; Porphyrin dendrimers; Poly (propyl) ether imine dendrimer; Zinc porphyrin dendrimer
1. Introduction
Dendrimers are highly branched, star-shaped macromolecules with nanometer-scale dimensions. Dendrimers are defined by three components: a central core, an interior dendritic structure (the branches), and an exterior surface with functional surface groups. Dendrimers have a huge range of applications in all branches of chemistry, especially in host–guest reactions and self-assembly procedures. Dendrimers are used in the formation of nanotubes, nanolatex, chemical sensors, micro/macro capsules, coloured glass, modified electrodes, and photon funnels such as artificial antennas [1-13]. Because dendrimers are widely used in different applied fields, the study of nanostar dendrimers has received a great deal of attention in both chemical and mathematical literature [13-24].
Molecules and molecular compounds are often modeled by molecular graphs. A molecular graph is a representation of the structural formula of a chemical compound in terms of graph theory, whose vertices correspond to the atoms of the compound and edges correspond to chemical bonds. A graph G (V, E) with vertex set V and edge set E is connected, if there exists a connection between any pair of vertices in G. For a graph G, the degree of a vertex v is the number of edges incident with v and denoted by deg (v).
A graph can be recognized by a numeric number, a polynomial, a drawing, a sequence of numbers, or a matrix. A topological index is a numeric quantity associated with a graph that characterizes the topology of the graph and is invariant under graph automorphism. Many topological indices are widely used for quantitative structure-property relationship (QSPR) and quantitative structureactivity relationship (QSAR) studies. Among various topological indices, degree based topological indices are the most important and widely used. These have great application in chemical graph theory. Since the 1970s, two degree based graph invariants have been extensively studied. These are the first Zagreb index M1 and the second Zagreb index M2, defined as

Details on the two Zagreb topological indices can be found in [2-6]. Randi c’ index was proposed by the chemist Randic [20] in 1975 and is defined as

The widely used connectivity topological index is atom-bond connectivity ( ABC ) index introduced by Estrada et al. [10]. The ABC index of graph G is defined as

Geometric-arithmetic (GA) index is another well known topological index. It was shown that its predictive power is better than the Randi `c index for many physicochemical properties like entropy, boiling point, vaporization, enthalpy, enthalpy of formation and acentric factor etc. This topological index is defined by Vuki c evi c [21] as follows:

For recent results on vertex-degree based topological indices, we refer Hua and Ning [22], Nadeem et al. [23,24]. Recently, Gutman et al. [10,13] re-introduced the neglected topological indices and succeed to demonstrate that these indices also have very promising applicative potential. The new/old topological indices studied by Gutman et al. [10,13] are the following: The reciprocal Randi  index (RR) is defined as (Figure 1).
index (RR) is defined as (Figure 1).

Obviously it is a special case of general Randi c’ index  where α is a real number. The reduced Randi c’ index is defined as
where α is a real number. The reduced Randi c’ index is defined as

The reduced second Zagreb index is defined as

2. Three New/Old Index of Poly (Propyl) Ether Imine Dendrimer
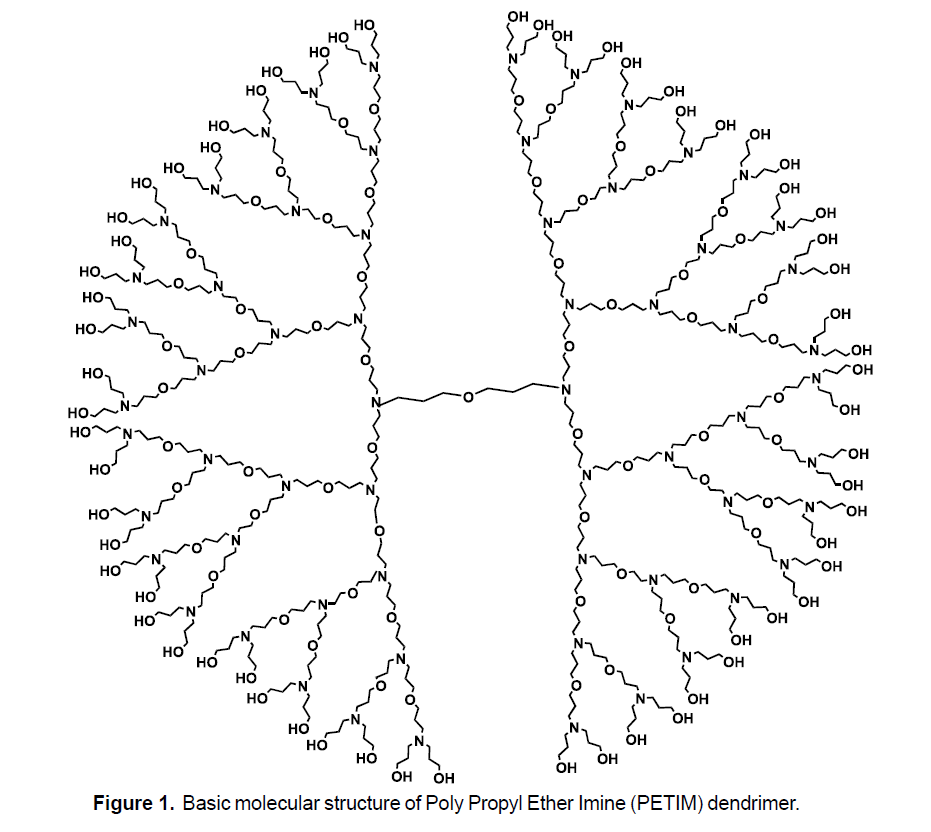
In this section, we study the RR, RRR and RM2 indices of PETIM dendrimer of generation with n growth stages. The molecular structure for the growth of PETIM dendrimer is shown in Figure 2. It is easy to see that the graph of PETIM dendrimer has 24×2n-23 vertices and 24×2n-24 edges. Now we compute RR, RRR and RM2 indices of PETIM dendrimer.
Theorem 1 Let G be the molecular graph of Poly(Propyl) Ether Imine(PETIM) dendrimer. Then

Proof. Let G be the graph of PETIM dendrimer. We have  and
and There are three partitions of edge set correspond to their degrees of end vertices which are:
There are three partitions of edge set correspond to their degrees of end vertices which are:

and

With the help of this partition we can easily find the required results. We apply these to the formulas of RR, RRR and RM2 to compute these indices for G. Since,


3. Three New/Old Index of Porphyrin Dendrimers
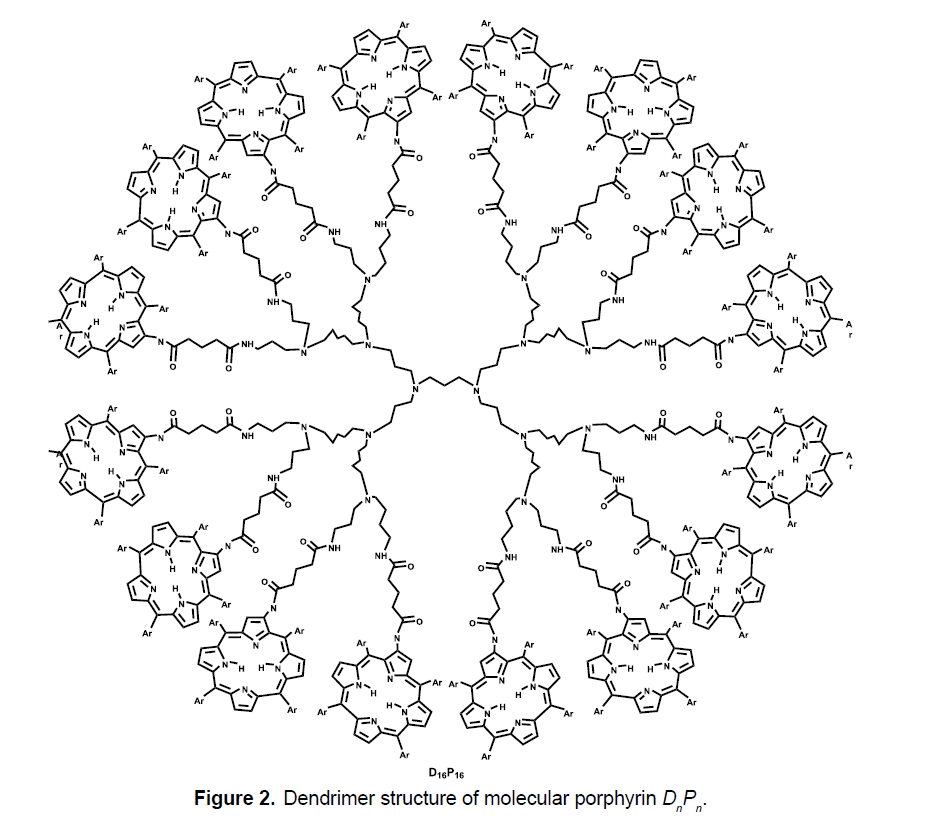
We consider the class of Porphyrin dendrimers, denoted by DnPn, where n is steps of growth. Note that n=2m, where m ≥ 2 (Figure 3). In the graph of DnPn, there are total 96n-10 vertices and 105n-11 edges. Figure 3 shows the graph of porphyrin dendrimer with growth stage n=3. Now we compute RR, RRR and RM2 indices of Porphyrin DnPn dendrimer.
Theorem 2 Let DnPn be a Porphyrin dendrimer. Then

Proof. Let G be the graph of DnPn dendrimer. We have  There are six partitions of edge set correspond to their degrees of end vertices which are
There are six partitions of edge set correspond to their degrees of end vertices which are


With the help of this partition we can easily find the required results. We apply these to the formulas of RR, RRR and RM2 to compute these indices for G. Since,


4. Three New/Old Index of Zinc-Porphyrin Dendrimer
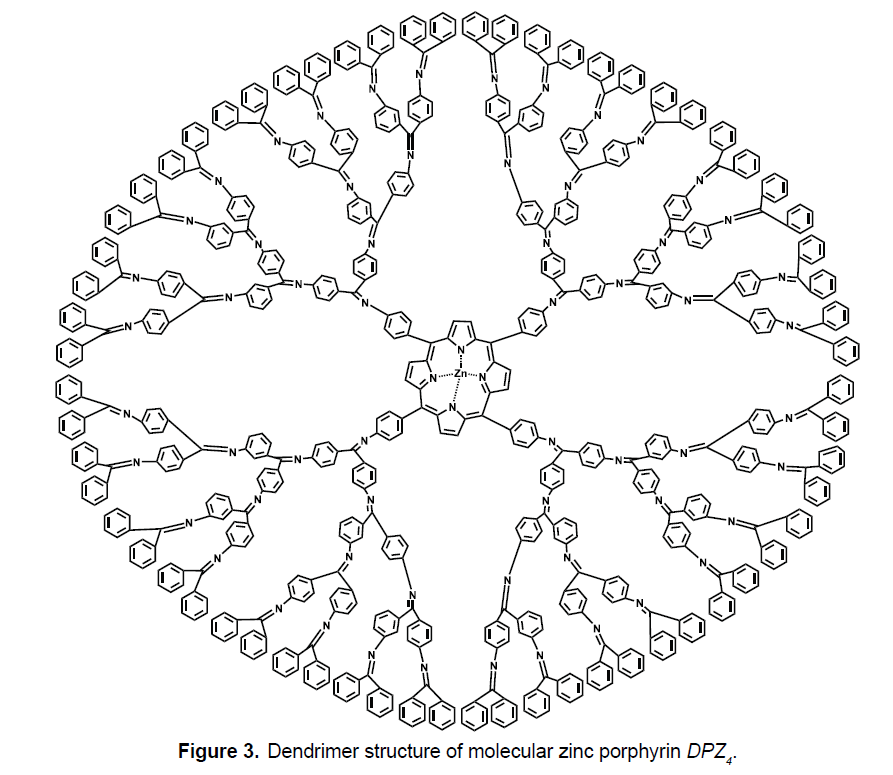
We consider the class of dendrimer Zinc-Porphyrin DPZn (Figure 3), where n is the steps of growth and n ≥ 1. In the molecular graph of DPZn there are total 56×2n-7 vertices and 64×2n-4 edges. Figure 3 shows the graph of Zinc-Porphyrin dendrimer with growth stage n=3. Now we compute RR, RRR and RM2 indices of Zinc-Porphyrin DPZn dendrimer.
Theorem 3 Let DPZn be a Zinc-Porphyrin dendrimer. Then

Proof. Let G be the graph of Zinc-Porphyrin DPZn dendrimer. We have |V (G) |= 56×2n − 7 and | E(G) |= 64×2n − 4 . There are three partitions of edge set correspond to their degrees of end vertices which are

and

With the help of this partition we can easily find the required results. We apply these to the formulas of RR, RRR and RM2 to compute these indices for G. Since,


5. Conclusion
In this paper we deal with three dendrimers families and studied their topological indices. We determined Randi c’ index (RR), the reduced second Zagreb index RM2, and the reduced reciprocal Randi c’ index (RRR) for these dendrimers families. Randi c’ index (RR) has proven its worth in so many drugs design and have been used at various occasions.
References
- Gutman I, Trinajstic N. (1972). Graph theory and molecular orbitals: Total p-electron energy of alternant hydrocarbons. Chem Phys Lett. 17: 535-538.
- Das KC, Gutman I. (2004). Some properties of the second Zagreb index, MATCH Commun. Math Comput Chem. 52: 103-112.
- Gutman I. (2014). On the origin of two degreebased topological indices. Bull Cl Sci Math Nat Sci Math. 146: 39-52.
- Gutman I, Das KC. (2004). The fi Zagreb index 30 years after, MATCH Commun. Math Comput Chem. 50: 83-92.
- Gutman I, Furtula B, Vukicevic ZK, et al. (2015). On Zagreb indices and coindices, MATCH Commun Math Comput Chem. 74: 516.
- Nikolic S, Kovacevic G, Milicevic A, et al. (2003). The Zagreb indices 30 years after, Croat. Chem Acta. 76: 113-124.
- Furtula B, Gutman I. (2015). A forgotten topological index. J Math Chem. 53: 1184-e1190.
- De N, Nayeem SMA, Pal A. (2016). F-index of some graph operations. Disc Math Alg Appl. 1: 18.
- De N, Nayeem SMA, Pal A. (2016). The F-co-index of some graph operations, Springer Plus. 5: 221.
- Estrada E, Torres L, Rodriguez L, et al. (1998). An atom-bond connectivity index: Modelling the enthalpy of formation of alkanes. Indian J Chem. 37: 849-855.
- Abdo H, Dimitrov D, Gutman I. (2015). On external trees with respect to the F-index. Discrete Mathematics. 1: 13.
- Diudea MV, Vizitiu AE, Mirzagar M, et al. (2010). Sad-hana polynomial in nano-dendrimers, Carpathian. J Math. 26: 59-66.
- Ashrafi AR, Mirzargar M. (2008). Szeged and edge Szeged of an infi family of nanostar dendrimers. Indian J Chem. 47: 538-541.
- Chen Z, Dehmer M, Emmert-Streib F, et al. (2014). Entropy bounds for dendrimers. Appl Math Comput. 242: 462-472.
- Diudea MV, Vizitiu AE, Mirzagar M, et al. (2010). Sad-hana polynomial innano-dendrimers, Carpathian. J Math. 26: 59-66.
- Diudea MV, Katona G. (1999). Molecular topology of dendrimers, In: G.A. Newkome (Ed.). Advan Dendritic Macromol. 4: 135-201.
- Ashrafi AR, Nikzad P. (2009). Connectivity index of the family of dendrimer nanostar. Dig J Nanomater Bios. 4: 269-273.
- Klajnert B, Bryszewska M. (2001). Dendrimers, properties and applications. Acta Biochim Pol. 48: 199-208.
- Yamamoto K, Higuchi M, Shiki S, et al. (2002). Stepwise radial complexation of imine groups in phenylazomethine dendrimers. Nature. 415: 509-511.
- Randic` M. (1975). On characterization of molecular branching. J Am Chem Soc. 97: 6609-6615.
- VukicÃÆââ¬Â¹Ãâââ¬Â¡evi`c D, Furtula B. (2009). Topological index based on the ratios of geometrical and arithmetic means of end-vertex degrees of edges. J Math Chem. 46: 1369-1376.
- Hua H, Ning B. (2017). Wiener Index, Harary Index and Hamiltonicity of Graphs, MATCH Commun. Math Comput Chem. 78: 153-162.
- Nadeem MF, Zafar S, Zahid Z. (2017). Some Topological Indices of L(S(CNCk[n])), Punjab University. J Math. 49: 13-17.
- Nadeem MF, Zafar S, Zahid Z. (2016). On the edge version of Geometric- Arithmetic index of nanocones, Studia universitatis babes. Bolyai Chemia. 61: 273-282.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences