PCR Restriction Fragment Length Polymorphism Analyses of V. Parahaemolyticus MAM-7 Virulence Gene in Clinical andEnvironmental Strains
Ana Elola-Lopez, Maria Jose Esquivel, Cristian A Munoz-Bergmann, Sebastian Beltran, Carlos G. Osorio, Annette N. Trombert
1Centro de Genomica y Bioinformatica,Facultad de Ciencias,Universidad Mayor,Santiago,Chile. Camino La Piramide 5750,Huechuraba
2Escuela de Tecnologia Medica,Facultad de Medicina,Universidad Mayor,Santiago,Chile. Camino La Pirámide 5750,Huechuraba
3Programa de Microbiologia y Micologia,Instituto de Ciencias biomédicas (ICBM),Facultad de Medicina,Universidad de Chile,Santiago,Chile. Avenida Independencia 1027,Independencia,Santiago,Chile
Received date: September 10,2015; Accepted date: October 20,2015; Published date: October 23,2015
Abstract
Virulent and non-virulent Vibrio parahaemolyticus (V. parahaemolyticus) strains coexist together in seawater. A PCR–restriction fragment length polymorphism (PCR-RFLP) technique could differentiate between clinical (virulent) and environmental V. parahaemolyticus strains. MAM- 7 corresponds to a virulence gene described in V. parahaemolyticus and that participates in initial stages of pathogen gut colonization. The objective of our study is to evaluate if PCR-RFLP analyses of MAM-7 gene can discriminate between clinical and environmental V. parahaemolyticus strains. Ten V. parahaemolyticus clinical isolates and nine V. parahaemolyticus environmental isolates were used to obtain genomic DNA. A 2619 bp PCR product from MAM-7 gene was digested with HindIII and AcuI restriction enzymes revealing a characteristic common pattern in 100% of V. parahaemolyticus clinical isolates. These patterns were absolutely different of those obtained from environmental isolates. PCR of toxin related genes (tdh and trh) showed that only clinical isolates were tdh+. As a conclusion, PCR-RFLP of V. parahaemolyticus MAM-7 gene could discriminate between clinical tdh+ isolates and environmental ones and could complement other diagnostic tools to detect and classify virulent strains. However, it is still necessary to analyze more samples of V. parahaemolyticus. Thus, while these results are promising, this study corresponds to preliminary work.
https://maviyolculuk.online/
https://mavitur.online/
https://marmaristeknekirala.com.tr
https://tekneturumarmaris.com.tr
https://bodrumteknekirala.com.tr
https://gocekteknekirala.com.tr
https://fethiyeteknekirala.com.tr
Keywords
Vibrio parahaemolyticus; RFLP; MAM-7.
Introduction
Gastrointestinal tract infections (GTIs) are one of the most common infectious diseases,affecting 1.7 billion people per year worldwide (WHO,2014). In addition,GTIs cause approximately 760.000 deaths per year in children under five years old (WHO,2014). GTIs are caused by diverse microorganisms such as viruses,parasites and bacteria that are found in contaminated water or food. Examples of bacteria that cause GTI are Escherichia coli,Clostridium difficile,Campylobacter,Salmonella,Vibrio among others [1]. Vibrio sp. grouped aquatic Gram negative bacteria and are commonly founded in marine and estuarine environment. From 12 pathogenic Vibrio species that infect humans,V. parahaemolyticus causes acute gastroenteritis (vibriosis) by consuming of undercooked seafood. Gastroenteritis is characterized by watery diarrhea,nausea,vomiting and abdominal pain [2,3].
The pandemic V. parahaemolyticus O3:K6 strain was first detected in Osaka (Japan) in 1950,and since 1996 this serotype has been spread throughout India,Europe,Africa,North,Central and South America [4,5]. Clinical V. parahaemolyticus strains isolated from human with vibriosis are usually differentiated from environmental strains by detection of tdh and trh genes and by their capability of hydrolyzing urea and inducing hemolysis in Wagatsuma blood agar (Kanagawa positive phenotype) [6,7].
Most environmental V. parahaemolyticus strains are considered non-virulent due to low detection of tdh and trh genes [7,8]. However,other studies show that most of V. parahaemolyticus strains carry one or more toxin genes [9]. In addition,there are strains that possess toxin genes and are Kanagawa negative [7]. Thus,phenotypic and genotypic parameters that are considered indicative of virulence in V. parahaemolyticus should be re-examined.
In this context,different molecular techniques have been designed to detect and classify V. parahaemolyticus strains such as PCR,Pulse Field Gel Electrophoresis (PFGE),ribotyping,Restriction Fragment Length Polymorphism (RFLP),multiplex PCR and PCR-RFLP [2,10,11]. PCR-RFLP and PFGE are methods used successfully to diagnose,discriminate and survey several food-borne bacterial pathogens. PFGE,considered the golden method in bacterial subtyping,implies obtaining of restriction patterns from whole bacterial genome that are analyzed and compared. However,PFGE is a timeconsuming and labour intensive method [12].
On the other hand,PCR-RFLP is a molecular technique characterized by the amplification of a nucleic acid sequence and its subsequent restriction enzyme digestion. Moreover,the presence of point mutations in the sequence amplified may alter the recognition sites of specific restriction enzymes. As a result,we can find differences in the restriction fragment profiles that can be compared between different strains [12-14]. Important advantages of PCR-RFLP include inexpensiveness,easily of experimental design,lack of requirement of advanced instruments and/or extensive training of laboratory staff [15]. Disadvantages include the requirements of specific (and sometimes expensive) restriction enzymes and difficulty to identify the variation in the nucleic acid sequence analyzed. Moreover,the analysis of several variations in different genes or sequences required similar number of specific primers and different restriction enzymes,limiting its usability for high throughput analysis [15]. However,RFLP analyses have been widely used for the identification of bacterial species and biotypes with excellent typeability,reproducibility,stability,and epidemiological concordance [12,13]. In fact,recently was observed the successfully use of RFLP to differentiate Salmonella biotypes [16],Escherichia coli enterohemolysin (ehxA) subtypes and pathogenic Vibrio species analyzing groEL gene [2].
Thus,we wanted to analyze the capacity of PCRRFLP to differentiate clinical and environmental V. parahaemolyticus strains analyzing the recently described adhesion factor,MAM-7 [17,18]. MAM-7 (Multivalent Adhesion Molecule 7) was conserved in several Gram negative bacteria and mediated the initial interaction between the pathogen and host cells. Because MAM-7 mediates cytotoxicity of V. parahaemolyticus,it is considered a new virulence factor [6,18]. Thus,in our study we employed a PCR-RFLP strategy to study MAM-7 gene as target in addition to the detection of tdh and trh genes to differentiate among strains obtained from clinical cases and those obtained from mussels (environmental isolates).
Methods
Bacterial strains and culture conditions
Reference strains V. parahaemolyticus [RIMD 2210633] serotype O3:K6 and V. parahaemolyticus [ATCC 17802] serotype O1:K1 were obtained from Instituto de Salud Pública (ISP),Chile. 10 strains serotype O3:K6 isolated from Chilean clinical cases of vibriosis (clinical strains,N° 149 - 701) and 9 strains isolated from mussels from Chilean coast (environmental strains,N° 3 -11),were obtained from Dr Carlos G Osorio collection (Instituto de Ciencias Biomedicas,Universidad de Chile,Chile). Strains were routinely growth in Luria-Bertani broth,supplemented with 1,5 % NaCl (LBS) (Merck) and cultured with agitation at 37°C for 12-24 h.
DNA extraction
Genomic DNA samples were obtained as described by Yue et al. [19] with some modifications. Briefly,a volume of 2 mL of bacterial cultures were centrifuged and bacterial sediments were resuspended in Tris- EDTA (pH 8.0) buffer. Each bacterial suspension was lysed using SDS 30 % and proteinase K (20 mg/mL) and incubated at 37°C for 1 hour. The lysates were mixed with guanidine thiocyanate solution (6M) and silica (50% p/v) and incubated 10 minutes at room temperature. Then,suspensions were centrifuged and supernatants were discarded. Silica from each sample was washed once with a wash buffer (guanidine thiocyanate 4,5M; Tris-HCl 50 mM; pH 7.2) and twice with ethanol 70%. Finally,each DNA sample was collected with nuclease-free water.
Amplification of the MAM-7 gene by PCR
Specific primers mam-7F (CGTATGTGCCTGATGTTAAGAGGA) and mam-7R (AAGGGCTTAGGAATTGGCGTT) were designed according available V. parahaemolyticus RIMD2210633 MAM-7 (VP1611) sequence (GenBank). PCR amplification reactions were performed using 1.25 U of Dream Taq® (Fermentas),60 ng of genomic DNA,0.2 μM of dNTP’s (Promega) and 1.5 mM of each primer. PCR conditions were: 1 cycle at 95°C for 5 minutes,followed by 30 cycles of denaturation (95°C for 30 seconds),annealing (65°C for 1 minute 30 seconds) and,extension (72°C for 2 minutes),with a final cycle at 72°C for 10 minutes. To verify the amplifications,PCR products were resolved in a 1% gel. According to the available sequence of V. parahaemolyticus MAM-7 (VP1611) gene,the expected PCR product size was of 2619 bp.
PCR-RFLP
PCR products of each V. parahaemolyticus strain were digested as follows: 0.3 μg of each PCR product was digested in a final volume of 20 μl with 10U of HindIII (Thermo Scientific) or 1 μl FastDigest AcuI (Thermo Scientific) at 37°C,according manufacturer’s instructions. Digestion products were resolved in a 2% agarose gel. Computational prediction of digestion patterns were performed using NEB Cutter 2.0,freely available in https://nc2. neb.com/NEBcutter2/ and RestrictionMapper (https:// www.restrictionmapper.org/).
PCR amplification of tdh and trh genes
Genomic DNA from 20 strains (from clinical and environmental strains) was used to amplify tdh and trh genes. Specific primer sequences were obtained from Tada et al [20]. Thus,TdhF (GGTACTAAATGGCTGACATC) and TdhR (CAACTACTCTCATATCG) were used for the amplification of tdh gene while TrhF (GGCTCAAAATGGTTAAGCG) and TrhR (CATTTCCGCTCTCATATAC) were used for the amplification of trh gene. PCR amplifications were performed using 1.25 U of Dream Taq® (Fermentas),60 ng of each genomic DNA,0.2 μM of dNTP’s (Promega) and 1.5 mM of each primer. PCR conditions were the following: 1 cycle of 95°C for 5 minutes,followed by 30 cycles of denaturation (95°C for 30 seconds),annealing (50.8°C for 30 seconds)and,extension (72°C for 2 minutes) with a final cycle at 72°C for 10 minutes. To verify the amplifications,PCR products were resolved in a 1% gel.
Results
Amplification of the MAM-7 gene
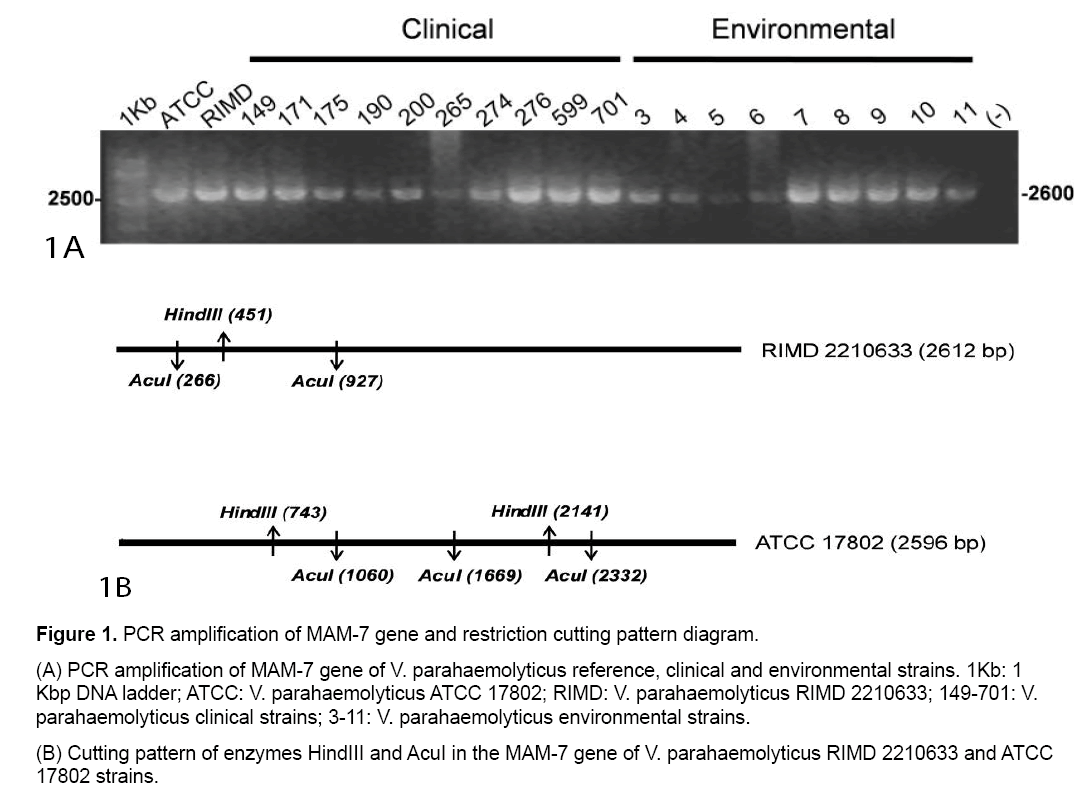
PCR amplifications from all clinical,environmental and reference V. parahaemolyticus strains yielded a PCR product of a size 2600 bp (Figure 1A).
Analysis and characterization of polymorphism of V. parahaemolyticus MAM-7 gene
Computational restriction fragment length analysis for V. parahaemolyticus RIMD 2210633 (RIMD) and V. parahaemolyticus ATCC 17802 (ATCC) MAM-7 PCR product using HindIII or AcuI restriction enzymes was performed. Schematic representations of digestion patterns for these reference strains are shown in Figure 1B and details of expected sizes in Table 1.
| Sample | Fragment sizes (bp) | |
|---|---|---|
| HindIII | AcuI | |
| V. parahaemolyticusRIMD2210633 | 450, 2162 | 265, 66, 1686 |
| V. parahaemolyticusATCC17802 | 456, 742, 1398 | 265, 609, 663, 1059 |
Table 1: Computational restriction fragment length analysis of the MAM-7 gene product obtained with the HindIII and AcuI enzymes.
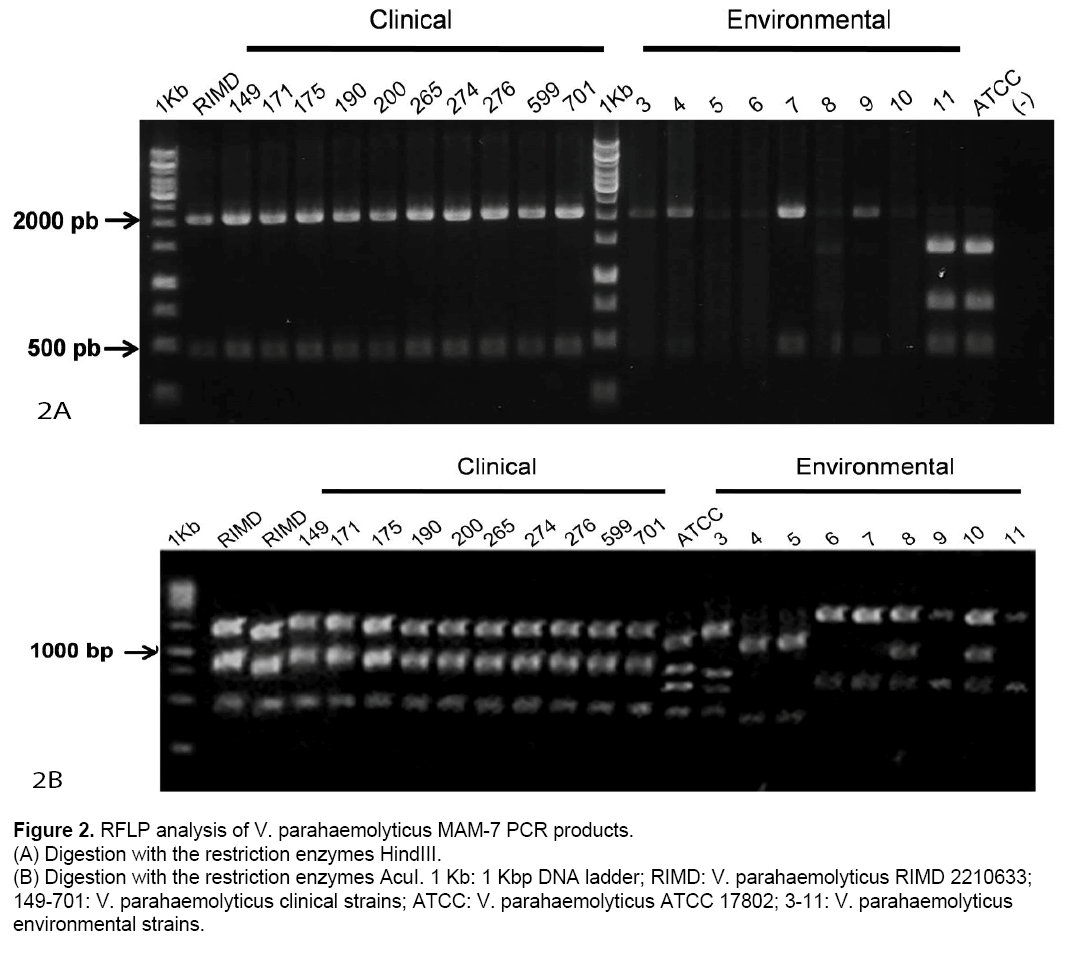
According these results,digestion patterns obtained from clinical strains are identical to those of RIMD using HindIII (450,2162 bp) as well as AcuI (265,661,1636 bp). Also,ATCC showed identical digestion patterns,as we predicted in silico (Table 1). In contrast,environmental strains showed digestion patterns that differ from those obtained from RIMD and ATCC using HindIII and/or AcuI (Figure 2 A and 2B).
Figure 2: RFLP analysis of V. parahaemolyticus MAM-7 PCR products.
(A) Digestion with the restriction enzymes HindIII.
(B) Digestion with the restriction enzymes AcuI. 1 Kb: 1 Kbp DNA ladder; RIMD: V. parahaemolyticus RIMD 2210633; 149-701: V. parahaemolyticus clinical strains; ATCC: V. parahaemolyticus ATCC 17802; 3-11: V. parahaemolyticus
environmental strains.
Detection of tdh and trh genes
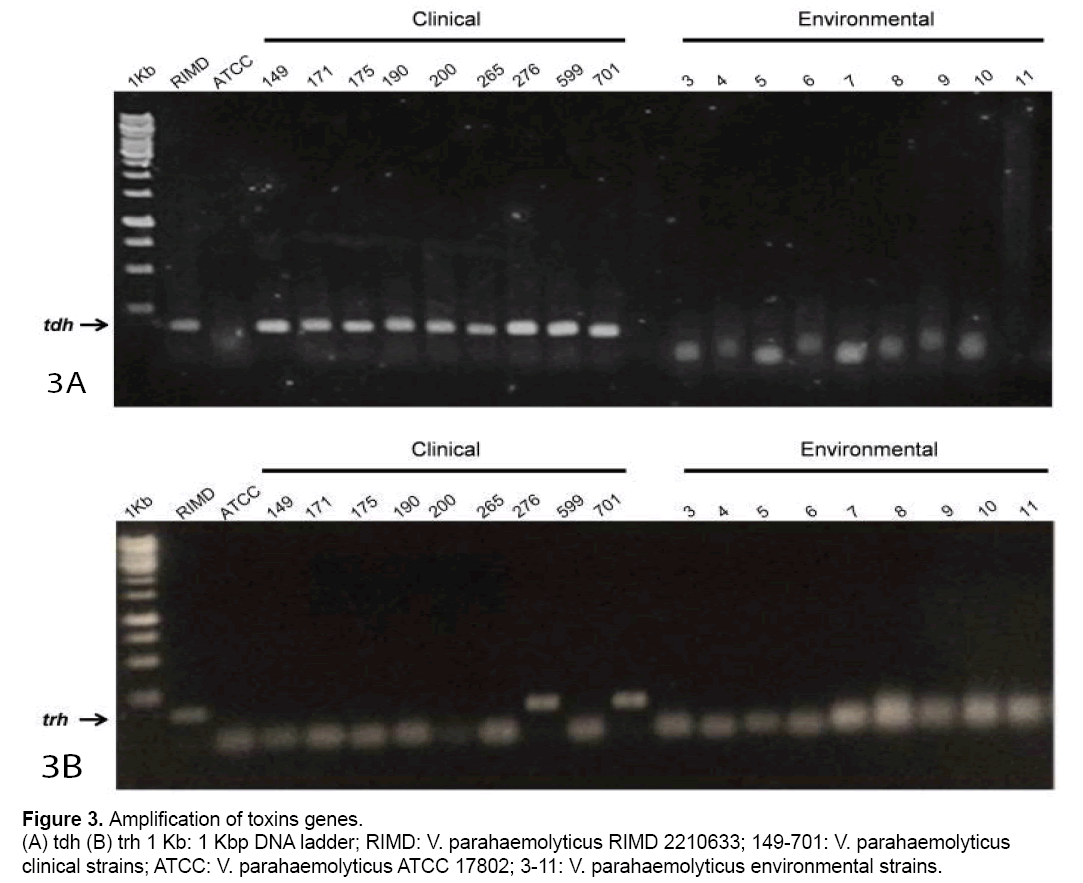
To establish a relationship between obtained digestion patterns and pathogenicity,toxins tdh and trh genes from clinical and environmental strains were amplified (Figure 3 A and 3B). Fig 3A showed that all clinical strains and RIMD amplified the trh gene. However,no amplification was observed in ATCC or environmental strains. Respect to the trh gene,it only was detected in RIMD and two clinical strains (Fig. 3B).
Discussion
In this study,we employed the PCR-RFLP-based strategy to screen for clinical and environmental-origin V. parahaemolyticus strains using MAM-7 gene as the target sequence. Simultaneously,we searched for tdh and/or trh genes using PCR in all strains studied. As a consequence,our study showed that MAM-7 gene is present in all V. parahaemolyticus strains analyzed. However,when we digested the PCR product of MAM-7 with restriction enzymes (PCRRFLP),a unique restriction pattern was showed in clinical pandemic strains and not in environmental strains. Coincidentally,only the clinical origin strains amplified for tdh toxin gene.
Traditionally,V. parahaemolyticus strains that possess tdh and trh genes are pathogenic for humans. Nevertheless,although infections produced by V. parahamolyticus are associated with mainly shellfish consumption,the number of isolates that possess trh and tdh genes varies greatly depending on the localization and detection techniques [21]. Thus,the development of new molecular approaches to differentiate pathogenic and non-pathogenic V. parahaemolyticus strains is necessary. PCRRFLP corresponds to a molecular technique that allowed differentiation of pathogenic strains where the traditional techniques as serotypification and conventional biochemical characterization have failed,or take several days to be performed [16,22].
PCR-RFLP was successfully used to identify Vibrio cholerae non-O1/non-O139,and to differentiate among pathogenic Vibrio species [2,23]. In our study,the V. parahaemolyticus MAM-7 gene was used as a target of PCR-RFLP. We studied 10 clinical and 9 environmental V. parahaemolyticus strains isolated from Chilean patients and mussels from Chilean coast. All clinical strains were tdh positive but only two of them (numbers 265 and 701) in addition to RIMD reference strain,amplified for trh. The previous results coincide with studies that highlight that not all clinical strains contain tdh and/or trh genes [24]. The absence of tdh PCR product in the reference strain ATCC 17802 correlated with previous antecedents [25]. On the other hand,no environmental strains amplified tdh and trh genes. The latter suggests that these strains lack two of the most relevant virulence factors. In relation with PCR-RFLP of MAM-7 gene,all clinical and RIMD reference strains,showed the same digestion pattern using Hind III and AcuI restriction enzymes. In relation to environmental samples,some strains showed a digestion pattern similar to the clinical strains with HindIII (strains numbers 3,4,7 and 9,Figure. 2A). However,these strains showed a completely different pattern with AcuI not only with respect RIMD but also with ATCC reference strains. Similar results were obtained with other restriction enzymes as SfcI,NspI,SfaN1 and HaeII (Data not shown). Thus,these results showed that V. parahaemolyticus MAM-7 gene is highly variable among strains and could be used as molecular marker. However,further evaluation of a higher sample size is necessary to verify this conclusion.
Recently,Hossain et al [2] showed that PCR-RFLP was more reliable than PCR-based method. In fact,PCR-RFLP-based strategy was used to screen for an array of pathogenic Vibrio species using groEL gene PCR product. In the study was shown that seven different human pathogenic species and two pathogenic species in fish presents unique digestion pattern [2]. In our study,our results showed that clinical V. parahaemolyticus O3:K6 strains presented unique digestion patterns compared with environmental and ATCC reference strains. V. parahaemolyticus strain ATCC 17802 correspond to an O1:K1 serovar isolated from food poisoning (www.ATCC.org) that also presented a digestion pattern different to the clinical and environmental strains isolated from mussels. Further evaluation of this new method using an array of clinical and environmental samples is necessary to evaluate if PCR-RFLP of MAM-7 could differentiate between serovars and/or strains of V. parahaemolyticus. Correlation between genotypes and clinical and/ or environmental origin does not necessarily mean that genotype analysis is diagnostic. In fact,genotype cannot predict unequivocally the virulence of an isolate [26]. Nevertheless,the identification of genes potentially useful as virulence markers,in combination with phenotypic analyses,can provide results that contribute to more precise diagnostic of infectious diseases.
Conclusions
PCR-RFLP of V. parahaemolyticus MAM-7 gene can be a potential complementary method to identify and characterize V. parahaemolyticus strains.
Acknowledgements
Escuela de Tecnología Médica,Facultad de Medicina,Universidad Mayor for restriction enzymes used for RFLP technique.
References
- Conlon,CP. (2010). Gastrointestinal Tract: Bacterial Infections. In Encyclopedia of Life Sciences (ELS). (John Wiley & Sons),Chischester.
- Hossain MT,Kim YR,Kong IS. (2014).PCR-restriction fragment length polymorphism analysis using groEL gene to differentiate pathogenic Vibrio species.DiagnMicrobiol Infect Dis.78: 9-11.
- Nair G,Ramamurthy T,Bhattacharya S,et al. (2007). Global Dissemination of Vibrio parahaemolyticus Serotype O3:K6 and Its Serovariants. Clin. Microbiol.20: 39–48.
- Velazquez-Roman J,León-Sicairos N,de Jesus Hernández-Díaz L,Canizalez-Roman A. (2014).PandemicVibrio parahaemolyticus O3:K6 onthe American continent. Front Cell Infect Microbiol.3: 110-14.
- Zhang L,Orth K. (2013). Virulence determinants for Vibrio parahaemolyticus infection. CurrOpinMicrobiol.16: 70-77.
- Letchumanan V,Chan KG,Lee LH. (2014).Vibrio parahaemolyticus: a review on the pathogenesis,prevalence,and advance molecular identification techniques. Front Microbiol.5: 705-718.
- Vieira RH,Costa RA,Menezes FG,Silva GC,et al. (2011).Kanagawa-negative,tdh- and trh-positive Vibrio parahaemolyticus isolated from fresh oysters marketed in Fortaleza,Brazil.CurrMicrobiol.63:126-130.
- Nishibuchi M,Kaper JB. (1995). Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium.Infect Immun.63:2093-2099.
- Gutierrez West CK,Klein SL,Lovell CR. (2013).High frequency of virulence factor genes tdh,trh,and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary.Appl Environ Microbiol.79:2247-2252.
- Hossain MT,Kim YO,Kong IS. (2013). Multiplex PCR for the detection and differentiation of Vibrio parahaemolyticus strains using the groEL,tdh and trh genes.Mol Cell Probes.27:171-175.
- Wang R,Zhong Y,Gu X,et al. (2015). The pathogenesis,detection,and prevention of Vibrio parahaemolyticus. Food Microbiol.6:1-13.
- Lukinmaa S,Nakari UM,Eklund M,Siitonen A. (2004).Application of molecular genetic methods in diagnostics and epidemiology of food-borne bacterial pathogens.APMIS.112:908-929.
- Al Dahouk S,Tomaso H,Prenger-Berninghoff E,et al. (2005).Identification of brucella species and biotypes using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP).Crit Rev Microbiol31:191-6.
- Çiftci IH,Ugras M,Acartürk G,et al.(2014) Comparison of FISH,RFLP and agar dilution methods for testing clarithromycin resistance of Helicobacter pylori.Turk J Gastroenterol25Suppl 1:75-80.
- Henrik Berg Rasmussen. (2012). Restriction Fragment Length Polymorphism Analysis of PCR-Amplified Fragments (PCR-RFLP) and Gel Electrophoresis - Valuable Tool for Genotyping and Genetic Fingerprinting. In Gel Electrophoresis - Principles and Basics (SamehMagdeldin),ISBN: 978-953-51-0458-2,InTech.
- Cheraghchi N,Khaki P,MoradiBidhendi S,Sabokbar A. (2014).Identification of Isolated Salmonella enterica Serotype gallinarum Biotype Pullorum and Gallinarum by PCR-RFLP.Jundishapur J Microbiol7:e19135.
- Lorenz SC,Fischer M,Kase JA. (2014).Improved PCR-RFLP method for the identification of Escherichia colienterohemolysin (ehxA) subtypes.J Microbiol Methods.100:24-6.
- Krachler AM,Ham H,Orth K. (2011). Outer membrane adhesion factor multivalent adhesion molecule 7 initiates host cell binding during infection by gram-negative pathogens. ProcNatlAcadSci USA.108:11614–11619.
- Yue GH,Orban L. (2001). Rapid isolation of DNA from fresh and preserved fish scales for polymerase chain reaction. Mar Biotechnol.3:199-204.
- Tada J,Ohashi T,Nishimura N,et al. (1992). Detection of the thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Molecular and Cellular Probes.6: 477-487.
- Yingkajorn M,Mitraparp-Arthorn P,Nuanualsuwan S,et al. (2014).Prevalence and quantification of pathogenic Vibrio parahaemolyticus during shrimp culture in Thailand.Dis Aquat Organ.112:103-111.
- Amhaz JM,Andrade A,Bando SY,et al. (2004). Molecular typing and phylogenetic analysis of enteroinvasiveEscherichia coli using the fliC gene sequence. FEMS MicrobiolLett.235:259-264.
- Awasthi SP,Asakura M,Neogi SB,et al. (2014).Development of a PCR-restriction fragment length polymorphism assay for detection and subtyping of cholix toxin variant genes of Vibrio cholerae.J Med Microbiol.63:667-673.
- Raghunath P. (2015). Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus.Front Microbiol.5:805-809.
- Cabrera-Garcia ME,Vazquez-Salinas C,Quiñones-Ramirez EI. (2004). Serologic and molecular characterization of Vibrio parahaemolyticus strains isolated from seawater and fish products of the Gulf of Mexico. Appl Environ Microbiol.70:6401-6406.
- Thiaville PC,Bourdage KL,Wright AC,et al. (2011). Genotype is correlated with but does not predict virulence of Vibrio vulnificus biotype 1 in subcutaneously inoculated,iron dextran-treated mice. Infect Immun.79:1194-1207.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences