Molecular Study of the Prevalence of CTX-M1, CTX-M2, CTXM3 in Pseudomonas aeruginosa Isolated from Clinical Samples in Tabriz Town, Iran
Abolfazl Jafari Sales, Reza Fathi, Haedeh Mobaiyen, Farnaz Rasi Bonab, Khatereh Babayi Kondlaji, Mobara Sadeghnezhadi
1Young Researchers and Elite Club, Ahar Branch, Islamic Azad University, Ahar, Iran
2Department of Physiology, School of Medicine, Marmara University, Istanbul, Turkey
3Department of Microbiology, Tabriz Branch, Islamic Azad University, Tabriz, Iran
4Young Researchers and Elite Club, Marand Branch, Islamic Azad University, Marand, Iran
5 PhD Student, Department of Microbiology, Kazeroon branch, Islamic Azad University, Kazeroon, Iran.
- *Corresponding Author:
- Abolfazl Jafari Sales
Young Researchers and Elite Club Ahar Branch
Islamic Azad University, Ahar, Iran
Tel: +98(0)914-7611841
Fax: +98(0)414-2274746
E-mail: a.jafari_1392@yahoo.com
Received date: May 28, 2017; Accepted date: June 29, 2017; Published date: July 06, 2017
Citation: Sales AJ, Fathi R, Mobaiyen H, et al. Molecular Study of the Prevalence of CTX-M1, CTX-M2, CTXM3 in Pseudomonas aeruginosa Isolated from Clinical Samples in Tabriz Town, Iran. Electronic J Biol, 13:3.
Abstract
Background: Pseudomonas aeruginosa is the most common pathogen causing nosocomial infections. One of the reasons for the drug resistance in the Pseudomonas aeroginosa strains is the production of Extended-Spectrum Beta-lactamases enzymes. This study aimed to determine the antibiotic resistance and the frequency of the Extended-Spectrum Betalactamases enzymes (CTX-M1, CTX-M2, CTX-M3) in the Pseudomonas aeroginosa strains isolated in the hospitals of Tabriz Town. Methods: The Pseudomonas aeroginosa strains were collected from different samples of patients admitted to hospitals and medical centers in Tabriz Town during the period from 26th December, 2014 to 25th December 2015. After identifying the phenotypic and genotypic identity (16Sr RNA) and performing antibiogram test, the phenotype of ESBLs in the bacterium of Pseudomonas aeruginosa was evaluated by double-disk synergy method. Then the bacterial DNA was extracted, studied and evaluated by PCR method and using special primers for the frequency of genes. Results: 110 Pseudomonas aeruginosa strains were identified from 1500 clinical specimens collected from the hospitals and medical centers in Tabriz Town that their highest resistance was related to the antibiotics of Amikacin (81.81%), Nalidixic acid (89.09%) and Ceftriaxone (75.45%) and the lowest one was related to the antibiotics of Tetracycline (44.54%) and Gentamicin (50.09%). The highest gene frequencies of ESBLs are related to the genes of CTX-M1 (27.27%), CTX-M2 (23.63%) and CTX-M3 (9.09%), respectively. Conclusion: The genes studied in this research were all on the chromosomes of the bacterium of Pseudomonas aeruginosa. So, investigating the genes of ESBL such as CTX-M1 which has the maximum frequency in the clinical specimens of Pseudomonas aeruginosa seems necessary.
Keywords
ESBL (Extended-Spectrum Betalactamases), gene of CTX-M, Pseudomonas aeruginosa
1. Introduction
Nosocomial infections are one of the most serious health problems in developed and developing countries [1]. Pseudomonas aeruginosa is the third nosocomial infection after Staphylococcus aureus and Escherichia coli [2]. Pseudomonas aeruginosa is one of the most important bacteria isolated from different clinical samples. Gram-negative bacteria, especially Pseudomonas aeruginosa, have intrinsic resistance to penicillin and the most of the antibiotics of beta-lactam but they are susceptible to the antibiotics of piperacillin, ciprofloxacin, tobramycin and imipenem [3]. This bacterium can be clearly seen in people who are immunosuppressed patients, for example the people who suffer burn, respiratory diseases, cancer (undergoing chemotheraphy), hereditary diseases of Cystic Fibrosis, bacteremia, septicemia and many other nosocomial infections [4,5]. Additionally, with the formation of biofilm, it creates the ability of protection against the host immune system and different antimicrobial agents [6,7]. Mortality in immunocompromised patients with pneumonia caused by Pseudomonas aeruginosa is about 79 percent [8]. Increasing antibiotic resistance has caused a lot of problems in patients and also in the treatment and increased the mortality [9]. One of the problems associated with Pseudomonas aeruginosa is multidrug resistance caused by different mechanisms, such as the production of cephalosporinase enzymes (due to the gene of AmpC) and beta-lactamase enzymes, reduced permeability of the outer membrane (reduction of protein) OprD, synthesis of the enzymes such as Phosphotransferase and Acetyltransferase (causing resistance to aminoglycosides) and also, the change in topoisomerase II and IV (resistance to quinolones) [10]. In addition to mentioned enzymes, there is another mechanism in the antibiotic resistance of Pseudomonas aeruginosa which is called efflux pump. Since this bacterium is of the bacteria which don’t need to grow more, it can easily remain in the environment and be transmitted to predisposed patients [11]. The high resistance of this bacterium to antimicrobial agents, including antibiotics, makes the treatment of the infections caused by this bacterium more complex and also, makes it as a major medical problem [12]. It seems that the survival of this bacterium in the environment and transmission of it to patients by various factors are the most important factor of the prevalence of these infections in these centers. In this regard, it has been tried to examine this problem by phenotypic and genotypic methods [13,14]. In the treatment of infectious diseases, determining antibiotic resistance, which should be done before starting the treatment, is very important. Also, indiscriminate use of antibiotics must be avoided to prevent the emergence of resistant strains [15]. Major Pseudomonas infections shouldn’t be treated just with one drug because the bacteria would quickly become resistant to it [16,17]. To treat these infections, a type of active penicillin such as Ticarcillin or Piperacillin is prescribed plus new generation of cephalosporins such as ceftazidime [16]. The aim of this study was to examine the betalactams resistance genes in clinical samples isolated from hospitals and medical centers in Tabriz Town.
2. Materials and Methods
2.1 Collection and identification of samples
This study is a descriptive-analytical research. During a year (26th December, 2014 to 25th December 2015), 1500 different clinical samples, including tracheal chip, blood, ulcer, sputum, fluids, urine, were collected from the patients admitted to medical centers, hospitals and specialized laboratories by randomly sampling method and also, the required characteristics of the patients, including gender, age and be inpatient or outpatient, were collected. In order to determine the phenotypic identity with the use of gram staining and to determine the form of microscopic organisms, oxidase and catalase tests and other biochemical tests, including Simmons Citrate test, production of H2S, production of indole and motility, reduction of nitrate, urease test, triple sugar iron agar, ornithine decarboxylase and lysine, DNase and oxidative-fermentative test were performed. For the final determination of the type, the molecular detection method of PCR (Polymerase Chain Reaction) was used with the gene of 16S rRNA (16S ribosomal RNA). For this purpose, all of samples were kept in the transitional environment of TBS (Tryptic Soy Broth).
2.2 Antibiogram test and double disk synergy to identify ESBL-producing strains
The antibiogram test was performed to investigate the drug resistance with the standard method of Kirby Baur and based on the standards of NCCLS and also to prepare 0.5 McFarland concentration of bacteria and to culture them in Mueller-Hinton agar medium and to investigate the resistance of isolated strains to the antibiotics of imipenem, ciprofloxacin, cefotaxime, ceftazidime, nalidixic acid, ceftriaxone, gentamicin, tetracycline, amikacin and ampicillin, made by PadtanTeb Co., at the plate level and according to the standards. Then, the phenotype identification of ESBL was done by using diffusion disk method and double disk synergy test and the use of ceftazidime and cefotaxime disks with and without amoxicillin/clavulanic acid and 0.5 McFarland concentrations of bacteria.
2.3 Identification of species based on 16Sr RNA gene
In this investigation, extraction of the genome of bacteria was performed by using extraction kit (InvitekStratec Business) (Made in Canada). Then, PCR test was done on the extracted gene for 16Sr RNA gene. In this study, two pairs of primers with the sequence of F:GGGGGATCTTCGGACCTCA and R:TCCTTAGAGTGCCCACCCG were used together in a PCR reaction to identify the species [18]. Required materials to do PCR reaction were: 1 μl of Bacterial Chromosom, 1μl of forwaed primer, 1 μl of revers primer, 0.5 μl of dNTP, 1 μl of MgCl2, 2.5 μl of 10x PCR buffer, 17.5 μl of D.W and 0.5 μl of Taq polymerase enzyme that its volume was increased to 25 μl and the thermal cycle with following steps were repeated for 30 times: primary denaturation at 95°C for 5 min, then, secondary denaturation at 95°C for 30 s, annealing at 58°C for 45 s, extension at 72°C for 1 min and finally, final extension at 72°C for 5 min. The strain of Pseudomonas aeruginosa PAO (ATCC 27853) was used as a positive control.
2.3 Identification of the genes of CTX-M1, CTX-M2, CTX-M 3
In the following, the bacteria whose ESBL phenotype was reported positive, were nightly cultured in LB (Luria-Bertani) liquid medium at 37°C for 24 h and then, DNA was extracted from the bacteria of Pseudomonas aeruginosa using InvitekStratec Business kit (made in Canada). Then, they were used on the genes of CTX-M1, CTX-M2 and CTX-M3 based on Table 1 with the specific oligonucleotide primers which were simultaneously used for the process of PCR (Multiplex PCR) [19]. Thermal cycler device was accurately set up for doing PCR process based on the mentioned resources in Table 2 [20]. SPSS V20 software and chi-square test were used to analyze the data statistically. The significance level was p<0.05.
| Primer Name | Amplicon Size | Nucleotide Sequence | References |
|---|---|---|---|
| CTX-M1 | 499 | F-GACGATGTCATCGGCTGAGC R-AGCCGCCGACGCTAATACA | 8 |
| CTX-M2 | 351 | F-GCGACCAGGTTAACTACAATCC R-CGGTAGTATTGCCCTTAAGCC | 8 |
| CTX-M3 | 307 | F-CGCTTTGCCATGTGCAGCACC R-GCTCAGTACGATCGAGCC | 8 |
Table 1. Primers used in this study.
| Test Procedure | Temperature (ÃÆââ¬Å¡ÃâðC) | Time (s) |
|---|---|---|
| Initial denaturation step | 94 | 180 |
| Denaturation step | 94 | 60 |
| Annealing step | 55 | 30 |
| Extension/elongation step | 72 | 60 |
| Final extension/elongation step | 72 | 600 |
Table 2. Steps and thermal cycle of the genes of CTX-M1, CTX-M2, CTX-M3.
4. Results
In this study, 1500 samples were collected from the hospitals and medical centers of Tabriz Town that 110 (7.3%) of them were the isolates of Pseudomonas aeruginosa. The highest frequency of Pseudomonas aeruginosa was isolated from Sina Hospital (Table 3). The mean age of patients was 37.4 ± 27 and variation range was between 10 months to 75 years old. 32 (29.09%) samples were related to men and 78 (70.91%) samples were related to women. Totally, the clinical samples included 29 (26.36%) tracheal chip samples, 25 (22.72%) blood samples, 19 (17.27%) ulcer samples, 14 (12.72%) sputum samples, 14 (12.72%) joint fluids samples and 9 (8.18%) urine samples. 70.9% of samples were collected from hospitalized sector and 29.1% of them were collected from outpatients. In terms of phenotype, 40 samples were positive that 25 (62.5%) of them were isolated from women and 15 (37.5%) of them were isolated from men.
| Health Centers/ Genus | Private Health Care Centers in Tabriz | Sina Hospital | Central Medical Laboratory of Tabriz | Imam Reza Hospital |
|---|---|---|---|---|
| Pseudomonas aeruginosa | 13 | 72 | 19 | 6 |
Table 3. Samples collected from the hospitals.
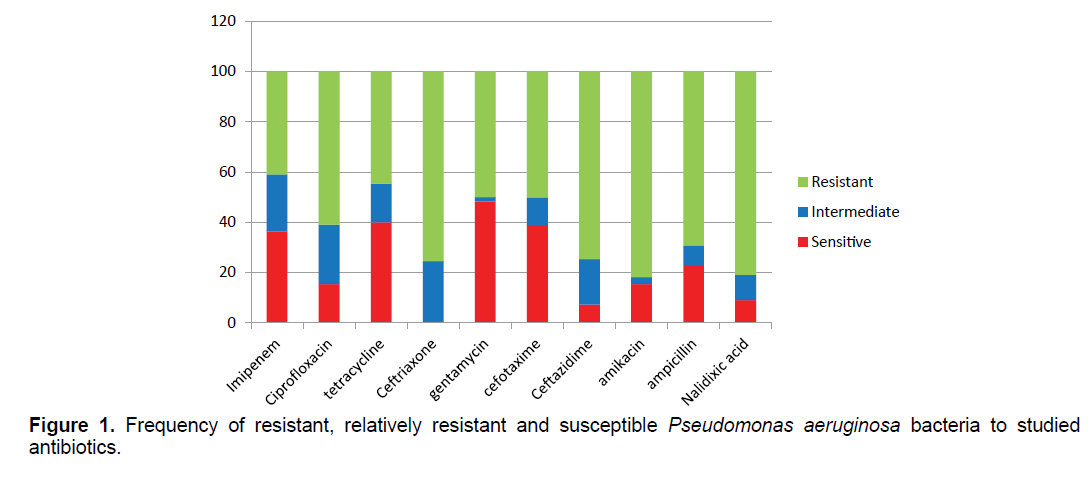
The results of antibiogram show that Pseudomonas aeruginosa bacteria have the highest resistance to the antibiotics of nalidixic acid (89.09%), amikacin (81.81%) and ceftriaxone (75.45%) and the lowest resistance to the antibiotics of tetracycline (44.54%) and gentamicin (50.09%) (Figure 1). 27/87% of samples were resistant to more than two antibiotic classes.
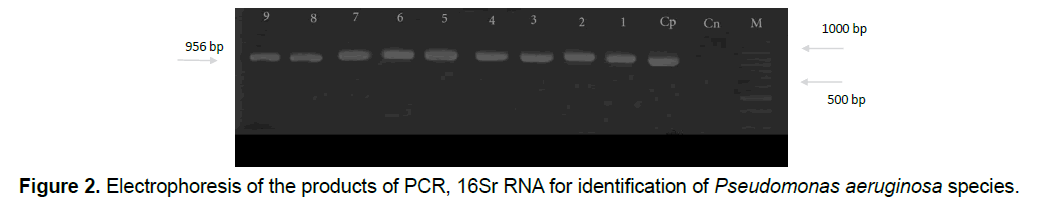
So it can be found that isolated Pseudomonas aeruginosa bacteria have the resistant genes resistant to mentioned antibiotics. After doing Polymerase Chain Reaction, in order to identify the species based on 16Sr RNA gene, according to the expected size of the species of Pseudomonas which is about 956 base pairs, the proliferation of Polymerase reaction with mentioned primers confirms a fragment of 956 base pairs that all samples of Pseudomonas (110 samples) were aeruginosa in terms of genotypic testing (Figure 2).
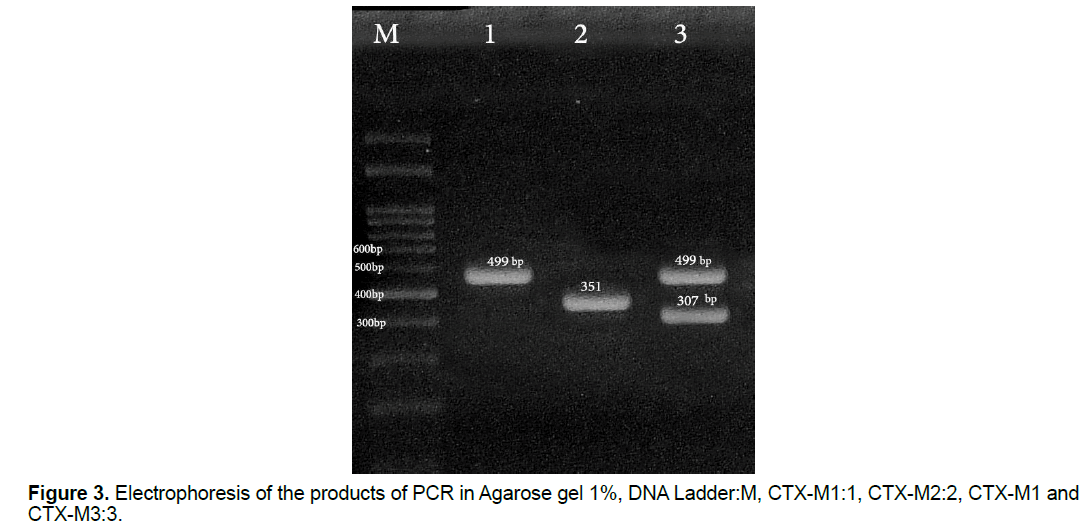
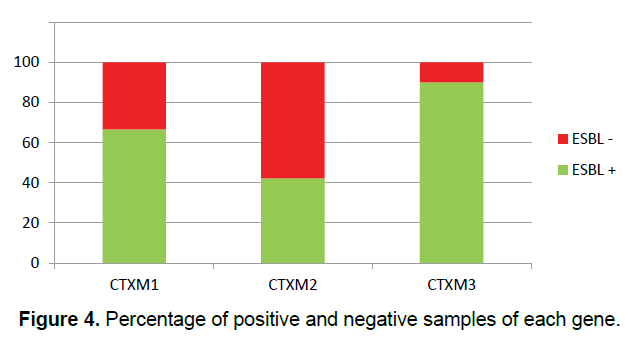
In this study, 30 (27.27%) samples of Pseudomonas aeruginosa bacteria have the gene of CTX-M1, 26 (23.63%) of them have the gene of CTX-M2 and 10 (9.09%) of them have the gene of CTX-M3 (Figure 3) and all of the Pseudomonas aeruginosa bacteria which their ESBL were positive in terms of the phenotype, have at least one of the studied genes of ESBL (Figure 4) and some of them have all three genes. This shows that all strains of isolated Pseudomonas bacteria have the studied genes of ESBL in their chromosomes, but, it is likely that some of the genes of ESBL are carried by the plasmid.
5. Discussion
The results showed that there was a significant relationship between the use of antipseudomonal drugs (amikacin, ciprofloxacin, ceftazidime, imipenem and etc.) and prevalence of resistant strains of Pseudomonas aeruginosa bacteria [21]. Also, in this study, the frequency of MDR Pseudomonas aeruginosa was high which is consistent with the results of other studies. The frequency of MDR in the isolates from burn patients has been reported 87.05% in Iran, 86.8% in Malaysia and 29.24% in Pakistan [22-24]. In this study, the highest prevalence of MDR was observed in the samples isolated from burn patients that show the high resistance of Pseudomonas aeruginosa to drugs used for burn wound infections [25].
In this study, the highest resistance was observed to the antibiotics of amikacin (81.81%), nalidixic acid (89.09%) and ceftriaxone (75.45%). Wesam et al. have examined the resistance pattern of Pseudomonas to the antibiotics and the results showed that this bacterium has the highest resistance to the antibiotics of nalidixic acid and tetracycline [26]. In another study in Arak in 2013, the resistance of this bacterium to the following antibiotics has been reported: ceftazidime (33.3%), imipenem (22.2%), amikacin (20.3%), ciprofloxacin (15.7%) and gentamicin (19.4%) [27]. In the study by Ahadi et al., the resistance of it to imipenem (55%) and ceftazidime (57%) has been reported [28]. Rakesh et al. [3] have reported the resistance of it to the antibiotics of ciprofloxacin (49%), gentamicin (63%) and imipenem (14%) in their study. Anli et al. [29] has reported the resistance of it to the antibiotics of amikacin (25%), ciprofloxacin (75%).
Kianpour et al. [30] have reported the resistance of it to the antibiotics of amikacin (58.14%), ciprofloxacin (42.58%) and imipenem (14.8%).
The results of the most of mentioned studies show the resistance of the isolates less than the one obtained in this study that the overuse and inappropriate prescription of antibiotics in recent years has been probably one of the reasons for the increased antibiotic resistance of this bacterium. Pseudomonas aeruginosa, due to its genetic nature, is the host of different genes through plasmids and transposons, maybe because of it, these bacteria can quickly become resistant to various antibiotics [31]. The production of ESBL in the isolates of Pseudomonas aeruginosa is on the rise in the last few years. The rate of ESBL increase was 20.6% in Thailand in 2003 [32], 25.4% in 2005 in Korea [33], 23.4% in 2006 in Bolivia and 45.3% in China in 2006 [34].
In the study by Mirsalehian et al., the production of ESBL in the clinical strains isolated in Tehran, has been reported 40% which is consistent with the results of present study [35].
In this study, the results of phenotype test on 110 isolates of Pseudomonas aeruginosa showed that the prevalence of Extended-Spectrum beta-lactamases producing strains was positive in 40 (36.36%) isolates, whereas the results of other studies are as following:
Shakibaie et al. [36] have been reported that 41 (34%) of 120 isolates of Pseudomonas aeruginosa were ESBL-producing strains. Shahcheraghi et al. [37] have been reported that 234 (39%) of 600 isolates of Pseudomonas aeruginosa were Extended-Spectrum beta-lactamases producing strains. These results show the higher frequency than the results of present study, this seems the presence of dominant resistant and ESBL-producing bacteria and treatment pattern associated with extended-spectrum cephalosporins cause the increase of ESBL-producing bacteria. In the study by Shahcheraghi et al. [38] the presence of the genes of VEB, OXA-10, CTX-M, PER-1, GES- 1, OXA-1, OXA-4 in the Pseudomonas aeruginosa strains isolated from the hospitals in Iran has been confirmed. In the studies performed in last years in Iran, the prevalence of ESBL enzymes, especially CTX-M, has been increased. In present study, the frequencies of beta-lactamase enzymes of CTX-M1, CTX-M2 and CTX-M3 are 27.27%, 23.63% and 9.09%, respectively. This explains the high resistance of isolates to the third and fourth generation of cephalosporins. Further studies carried out in Iran, have reported different results for the frequency of CTX-M. In the study conducted in Kashan, the frequencies of CTX-M1, CTX-M2 and CTX-M3 have been reported 1%, 0.0% and 0.0%, respectively [39]. But in the study performed in Shiraz, the frequencies of CTX-M1, CTX-M2 and CTX-M3 have been reported 49.9%, 135%, 23.1%, respectively [40]. This difference may be due to the types of samples. In other countries, different results have been reported for the prevalence of ESBL. For example, based on the studies performed in Brazil (2010), the highest prevalence of the enzymes of ESBL was related to CTX-M2 (19.6%) [41]. In the studies performed in France (2010) and Japan (2003), the frequency of the enzyme of CTX-M in the Pseudomonas aeruginosa strains has been reported 0.0% that is probably due to the appropriate use of beta-lactam antibiotics, especially cephalosporins in these countries [42,43].
According to the study conducted in India in 2014, the prevalence of CTX-M1in the Pseudomonas aeruginosa strains has been reported 6.3% [44].
6. Conclusion
The results of this study imply on the high resistance of Pseudomonas aeruginosa strains to the antibiotics. In Iran, due to the indiscriminate use of antibiotics, particularly penicillin family, antibiotic resistance is growing rapidly and the patients have increased and spread this resistance because they haven’t completed their course of treatment, but, in other studies performed in other countries, the number of isolated resistant bacteria have been reported less than it although the sampling has been done more broadly.
7. Acknowledgement
We would like to thank the Department of Microbiology, Islamic Azad University Marand and Tabriz for their laboratory equipment and technical support.
References
- Hepburn AL, Narat S, Mason JC. (2010). The management of peripheral blood cytopenias in systemic lupus erythematosus. Rheumatol. 49: 2243-2254.
- Bartels CM and Ramsey-Goldman R. (2014). Updates in US systemic lupus erythematosus epidemiology: Tales of two cities. Arthritis Rheumatol. 66. 242-245.
- Amaylia OE, Hendarsyah S, Sumartini D, et al. (2013). The Role of Neutrophil Lymphocyte Count Ratio as an Inflammatory Marker in Systemic Lupus Erythematosus, Acta Medica Indonesiana. The Indonesian Journal of Internal Medicine.170-174.
- Perhimpunan Rheumatology Indonesia. (2011). Rekomendasi diagnostic and prognostic systemic lupus erythematosus. Acta Medica Indonesiana. The Indonesian Journal of Internal Medicine. 170-174.
- Celikbilek M, Dogan S, Ozbakir O, et al. (2013). Neutrophil-lymphocyte ratio as a predictor of disease severity in ulcerative colitis. J Clin Lab Anal. 27: 72ÃÆâÃâââ¬Ãâââ¬Å76.
- Turkmen K, Erdur FM, Ozcicek F, et al. (2013). Platelet-to-lymphocyte ratio better predicts inflammation than neutrophil-to-lymphocyte ratio in end-stage renal disease patients. Hemodial Int. 17: 391-396.
- Bertsias GK, Tektonidou M, Amoura Z, et al. (2012). Joint European League against Rheumatism and European Renal Association European Dialysis and Transplant Association (EULAR/ERA- EDTA) recommendations for the management of adult and paediatric lupus nephritis. Ann Rheum Dis. 71: 1771ÃÆâÃâââ¬Ãâââ¬Å1782.
- Proctor MJ, Morrison DS, Talwar D, et al. (2011). A comparison of inflammation-based prognostic scores in patients with cancer. A Glasgow Inflammation Outcome Study. Eur J Cancer. 47: 2633-2641.
- Ginzler E, Tayar J. (2015). Fast Facts of Lupus. American College of Rheumatology Committee on Communications and Marketing.
- Lixiu L, Yuncheng X, Chunmei C, et al. (2015). Neutrophil-lymphocyte ratio in systemic lupus erythematosus disease: A retrospective study. Int J Clin Exp Med. 8. 11026-11031.
- Chua W, Charles KA, Baracos VE, et al. (2011). Neutrophil-lymphocyte ratio predicts chemotherapy outcomes in patients with advanced colorectal cancer. Br J Cancer. 104. 1288-1295.
- Baodong Q, Ning M, Qingqin T, et al (2016). Neutrophil-lymphocyte ratio and platelet -lymphocyte ratio were useful markers in assessment of inflammatory response and disease activity in SLE patients: Modern Rheumatology. 6: 372-376.
- Yunxiu W, Yanjuan C, Yihua Y, et al. (2016). neutrophil-lymphocyte ratio and platelet-lymphocyte ratio were associated with disease activity in patients with SLE. International Immunopharmacology. 36: 94-99.
- Delgado G, Galarza D, Colunga P, et al. (2015). Neutrophil lymphocyte ratio is not superior to lymphocyte count alone in SLE. Eur J Gastroenterol Hepatol. 27. 108.
- Akkaya E, Gul M, Ugur M, et al. (2014). Platelet to lymphocyte ratio: A simple and valuable prognostic marker for acute coronary syndrome. Int J Cardiol 28:13-15.
- Gorenwold J, Bronsveld W. (1986). Predictive value of complement profiles and anti-dsDNA in systemic lupus erythematosus. Ann Rheum Dis. 45: 359-366.
- Nived O, Sturfelt G. (2004). ACR classification criteria for systemic lupus erythematosus: complement components. Lupus. 13: 1-3.
- Stoll T, Seifert B, Isenberg DA. (1996). SLICC/ACR damage index is valid, and renal and pulmonary organ scores are predictors of severe outcome in patients with SLE. Br J Rheumatol. 32: 248-254.
- Haq I, Isenberg DA. (2002). How does one assess and monitor patients with systemic lupus erythematosus in daily clinical practice? Best Pract Res Clin Rheumatol. 16: 181-194.
- Jennings I, Kitchen S, Woods TA, et al. (1997). Potentially clinically important inaccuracies in testing for the lupus anticoagulant: an analysis of results from three surveys of the UK national external quality assessment scheme (NEQAS) for blood coagulation. Thromb Haemost. 77: 934-937.
- Mok CC, Lau CS. (2003). Pathogenesis of systemic lupus erythematosus. J Clin Pathol. 56: 481-490.
- Zein N, Ganuza C and Kushner I. (1979). Significance of C-Reactiveprotein elevation in patients with SLE. Arthritis Rheum. 22: 7ÃÆâÃâââ¬Ãâââ¬Å12.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences