Molecular Identification and Phylogenetic Affinity of Two Growth Promoting Fungal Endophytes of Sweet Potato (Ipomea batatas (L.) Lam.) from Baguio City, Philippines
Roland M. Hipol
Department of Biology, College of Science, University of the Philippines Baguio, Baguio City, Philippines
- Corresponding Author:
- Tel: (6374)442-7231, +639062104303
E-mail: rmhipol@gmail.com.
Abstract
A total of 36 fungal endophytes were isolated from apparently healthy sweet potato plants from leaves, stems and roots collected from Baguio City. Among the isolates, only P3AL2c (GenBank accession no. JN672600.1) and P3BS1c (GenBank accession no. JN672605) significantly enhanced growth of paclobutrazol treated rice seedlings. The genomic DNA of these two fungal endophytes were isolated and ran on PCR using ITS 1 and ITS 4 primers. The PCR products were sequenced and subjected to BLASTn for detection of homology to Genbank accessions. P3AL2c was closest to Fusarium oxysporum isolate UOA/HCPF and P3BS1c was most homologous with Emericella nidulans strain FH5. Phylogenetic analysis using maximum likelihood algorithm with 1000 bootstrap replications generated the phylogenetic affinities of the isolates with Fusarium/Gibberella clade and Emericella nidulans, respectively.
Keywords
fungal endophyte; growth promotion; ITS; phylogenetic affinity.
1. Introduction
The term endophyte was coined by the German scientist Heinrich Anton de Bary in 1884 [1] and defined these as fungi living inside plants. This vague and encompassing definition however created confusions on how this term is to be used. Currently, Arnold [2] defines fungal endophytes as “a polyphyletic group of highly diverse, primarily ascomycetous fungi defined functionally by their occurrence within asymptomatic tissues of plants, are found in above-ground tissues of liverworts, hornworts, mosses, lycophytes, equisetopsids, ferns, and seed plants from the arctic to the tropics, and from agricultural fields to the most biotically diverse tropical forests”.
Ecologically, fungal endophytes have been discovered to have different roles. They confer tolerance to several abiotic stresses such as drought [3] and salt stress [4]. They also confer adaptive traits against insect pests [5] and disease [6]. Growth promotion has also been observed to be one of the agriculturally important traits of fungal endophytes [7]. These often-overlooked ecological importances have inspired growing enthusiasm regarding these little known fungi over the past four decades [2].
Sweet potato (Ipomea batatas (L.) Lam.) is one of the important agricultural crops, especially in the Cordillera highlands of the Philippines. They are grown near households and at forest edges. These hardy plants are grown without any form of fertilizer relying on the plant’s resilience against biotic and abiotic stressors.
In this work, the endophytic mycobiota that has the capacity to improve growth in rice that naturally occur in Ipomea batatas found in Baguio City was investigated. Information about the systemic investigation of fungal community associated with all plant tissues of this species has not been done before. The assay for the fungal isolates’ growth promoting activity may help us understand their potential beneficial activity to sweet potato host plants and to suggest possible applications.
2. Materials and Methods
Isolation of the fungi
Six healthy looking Ipomea batatas (L.) Lam. plants with their leaves, stems and roots were collected from Baguio City. The collected plants were brought to the Ecology laboratory of the University of the Philippines Los Baños in ice and processed immediately. They were washed with tap water and representative leaves, stems and roots were excised. A 0.5 x 0.5 cm portion of the leaves and 0.5 cm segments of the stems and roots were surfaced sterilized following the protocol used by Gotz et al. [8]. Epiphytic fungi were eliminated in this step to ensure that only endophytes remain. The effectiveness of surface sterilization method was tested with imprints of the tissues in malt extract agar (MEA) plates. These segments were then incubated in MEA with Streptomycin to inhibit bacterial growth, and Rose Bengal to slow down the growth of fungi especially the vigorous ones. The plates were incubated for 60 days. Each fungal colony that grew was transferred into MEA slants.
Bioassay for growth promoting metabolites on IR 64 rice
The protocol used in this section followed that of Hamayun et al. [7] however modified to suit specific limitations of the laboratory. Mycelia was obtained from the isolates in MEA slants and grown in malt extract (ME) broth supplemented with 200mg of streptomycin to limit bacterial growth. After 2 weeks, the culture broth was filtered to remove the mycelia from the filtrate. The culture filtrate of the fungal isolates was assayed on IR 64 rice sprouts in order to identify the presence of plant growth promoting metabolites.
The seeds of IR 64 were surface sterilized with 10% NaOCl and treated with 20ppm of paclobutrazol (a GA inhibitor) for 24 h, in order to inhibit GA synthesis. The treated seeds were washed thoroughly and soaked in distilled water for germination. After 48 h the germinated seeds were transplanted in glass tubes containing a 0.8% water-agar medium and kept in growth chamber. At the two leafed stage, 50μl of the fungal culture filtrate was applied to the plants at their apices. The shoot lengths were observed 7 days after the application and compared with the growth of rice seedlings treated with sterile ME broth (- control) and 20 ppm GA (+ control) in ME broth. Each of the treatments and both the controls were all set up in triplicates. The values were subjected to Analysis of Variance to determine if there are significant differences between the measured shoot lengths using SPSS® version 17.0. The Least Significant Difference (LSD) post-hoc test of the same software was used to show the different homogeneous subsets that are significantly different from each other.
Genomic DNA extraction identification of growth promoting isolates
The mycelia of the fungal isolates from the broth culture were comminuted with the use of a micro-pestle in a microcentrifuge tube. The succeeding steps followed the protocol of the Vivantis GF-1® plant DNA extraction kit as per the manufacturer’s instructions. The isolated DNA was suspended in 150μl of the elution buffer and tested of its purity in 1% agarose gel.
Molecular identification
The endophytic fungal isolates were identified by sequencing the internal transcribed region (ITS) of the 18S rDNA, using universal primers ITS-1 (5’TCC GTA GAA CCT GCG G-3’) for the forward primer and ITS-4 (TCC TCC GCT TAT TGA TAT GC’) for the reverse primer. The PCR mixture (36μl) contained 12.5 μl of the Vivantis PCR Master Mix, 0.75 μl MgCl2, 1l each of the forward and reverse primers, 14.75 μl of nuclease free water, and 6 μl of the isolated DNA. The reaction cycle consisted of a pre-denaturation phase of 5mins at 95°C, followed by 35 cycles of 30s at 95°C, 1min at 60°C and 1min at 72°C. Final extension phase of 6mins at 72°C was also done. The PCR products were run on 1% agarose gel to check the amplification of the desired length which is about 550 bp. The PCR products were sent to Macrogen Inc. of Korea for cleaning and subsequent sequencing.
The Basic Local Alignment Sequence Tool for nucleotides (BLASTn) search program [9] was used to look for nucleotide sequence homology for the 18s, ITS (1/4) and 5.8s regions for the fungal isolates for their identification. Isolates whose sequences had a similarity greater than 95% were considered to belong to the same species. This 5% difference used to define species boundaries appears to correlate well with differences among known endophytic species [10]. In instances where the similarity is below 95%, only the genus was accepted.
Phylogenetic analysis
DNA sequences of the isolates together with downloaded fungal sequences from the Genebank were subjected to Multiple Sequence Alignment algorithm using ClustalX software. The alignment was exported to Molecular Evolutionary Genetics Analysis software version 5 (MEGA 5) for the editing of the sequences and consequent re-alignment of the sequences. MEGA 5 was also used to determine the most appropriate model for the Maximum Likelihood algorithm for the derivation of the phylogenetic affinities of the studied sequences. Maximum Likelihood analysis used 1000 bootstrap replications to come up with the consensus phylogenetic tree.
3. Results
3.1 Fungal endophyte isolation and test for growth promoting activity
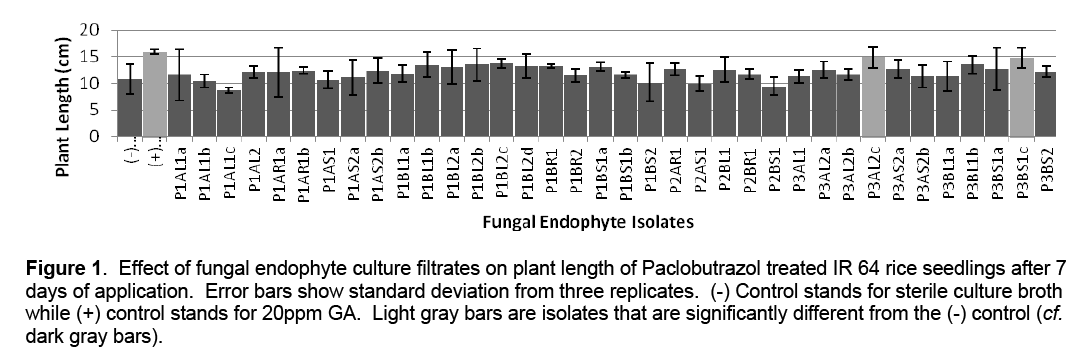
A total of 36 fungal endophyte pure cultures were isolated from apparently healthy Ipomea batatas plant tissues (leaves, stems and roots). These were transferred to MEA slants for temporary storage. For the growth promotion assay, the isolates were grown on MEA broth. The resulting fungal culture filtrates were tested on Paclobutrazol treated IR 64 rice varieties. Results show that that thirty isolates promoted plant growth with two isolates (P3AL2c and P3BS1c) being significantly different from the negative control (Figure 1).
Figure 1: Effect of fungal endophyte culture filtrates on plant length of Paclobutrazol treated IR 64 rice seedlings after 7 days of application. Error bars show standard deviation from three replicates. (-) Control stands for sterile culture broth while (+) control stands for 20ppm GA. Light gray bars are isolates that are significantly different from the (-) control (cf. dark gray bars).
3.2 Genomic identification and phylogenetic analysis of growth promoting isolates
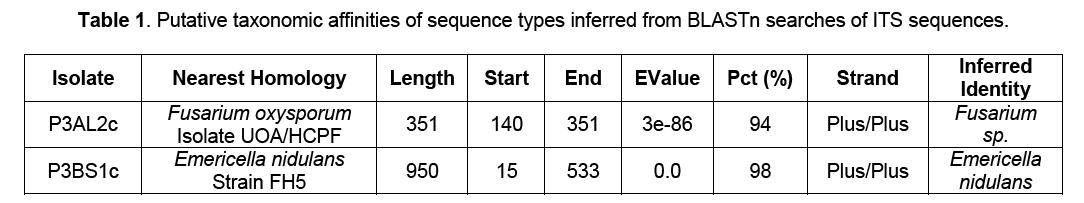
These two isolates were subjected to genomic DNA isolation and PCR targeting the adjacent sequences of the 18S ribosomal RNA gene, internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, and 28S ribosomal RNA gene using the ITS 1/4 primers. BLASTn search (Table 1) showed that P3AL2c closely resembled Fusarium oxysporum isolate UOA/HCPF with a 94% homology and P3BS1c was closest to Emericella nidulans strain FH5 with a homology of 98%. Following Arnold and Lutzoni’s [10] suggested threshold cut off for genus and species acceptance where 95% sequence similarity for the ITSrDNA region is designated as a conservative species boundary, it is inferred that the isolates are Fusarium sp. and Emericella nidulans respectively.
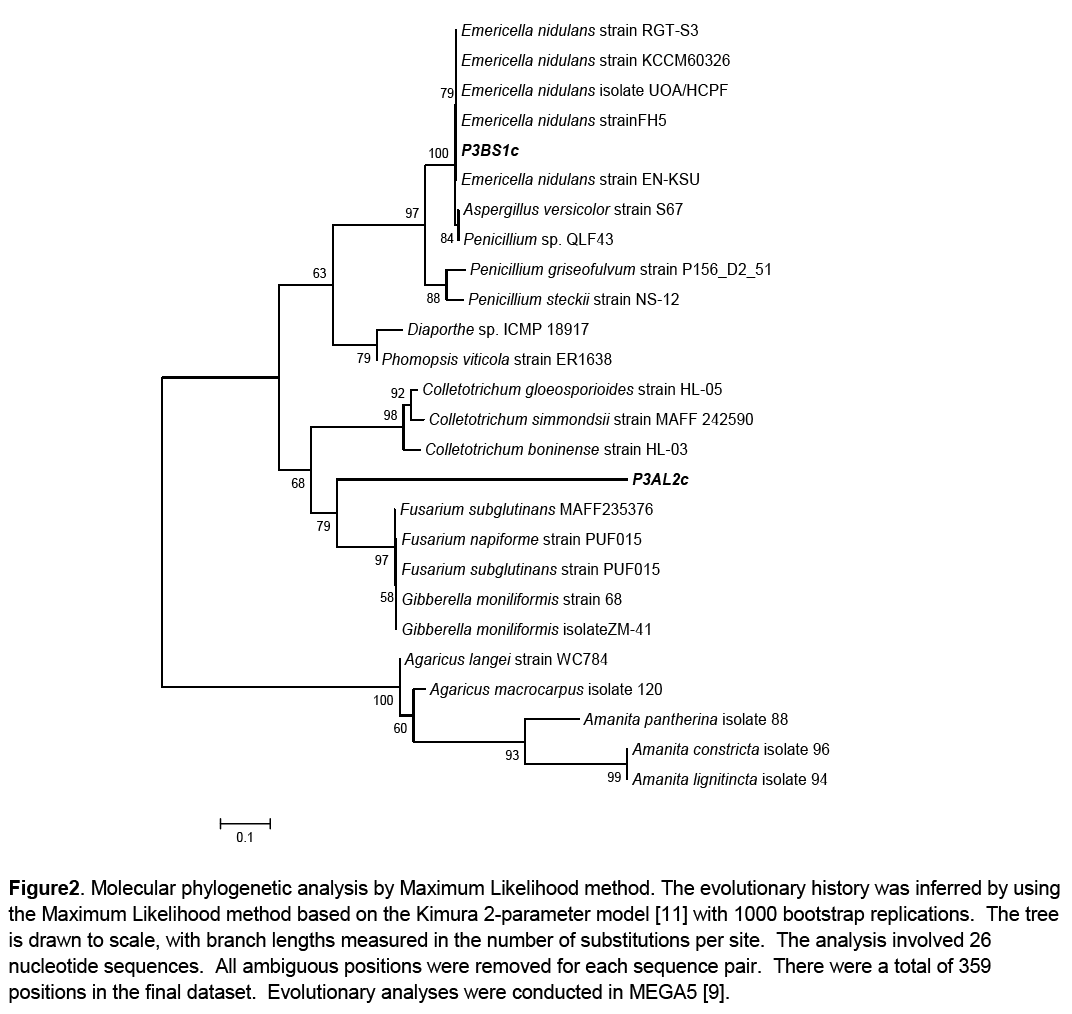
Phylogenetic analysis of the growth promoting fungal isolates was done to infer their phylogenetic affinities. This was carried out using the Molecular Evolutionary Genetics Analysis software version 5. The evolutionary history was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model [11]. A discrete Gamma distribution was used to model evolutionary rate differences among sites. The consensus tree was constructed from 26 aligned sequences (24 reference sequences and 2 clones) with 1000 bootstrap replications [12]. The reference sequences were selected through BLASTn using those that had the highest sequence percentage homology to the queries. Basidiomycete representative species of Agaricus and Amanita were used as an outgroup (Figure 2).
Figure 2: Molecular phylogenetic analysis by Maximum Likelihood method. The evolutionary history was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model [11] with 1000 bootstrap replications. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 26 nucleotide sequences. All ambiguous positions were removed for each sequence pair. There were a total of 359 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 [9].
4 Discussion
One of the identified primary objectives for the current researches on sweet potato in developing countries, including the Philippines, is the development of improved cultivars exhibiting high and stable yield potential [13].
Basic cultivar genotyping and breeding efforts can provide solution for this problem. Complementing such efforts is tapping ecologically important microorganisms such as fungal endophytes. These organisms are known to improve plant performance and productivity by increasing their host’s tolerance to biotic and abiotic stress and promoting growth.
Treatment of the IR 64 seeds with paclobutrazol, a GA biosynthesis inhibitor, suppresses the endogenous GAs production by blocking its biosynthesis pathway in the plant [14]. Also, the growth media were devoid of nutrients, it being water agar only. As such, growth promotion in the test seedlings can be attributed to the activity of plant growth promoting secondary metabolites from fungal culture filtrates. In this research therefore, it can be concluded that the significant increase in plant length for the seedlings treated with the culture filtrates of P3AL2c and P3BS1c were due to the presence of growth promoting metabolites from these fungal endophytes.
Several authors have also confirmed that fungal endophytes do produce growth promoting phytohormones. Khan et al. [15] have discovered that endophytic Paecilomyces formosus produce gibberellins and indole acetic acid. Hassan [16] also reported that Aspergillus flavus, A. niger, Fusar- ium oxysporum, Penicillium corylophilum, P. cyclopium, P. funiculosum and Rhizopus stolonifer have the capacity to produce GAs, while F. oxysporum can secrete both GAs and IAA.
Endophytic fungi secreting plant growth regulating compounds are of great agronomic importance to enhance crop yield and quality [15]. These growth regulating compounds can affect plant development as well as support plant growth in instances of biotic and abiotic stress such as tolerance to herbivory, heat, salt, disease, and drought, and increased below and above ground biomass [17].
This research provides helpful information on the possible role of fungal endophytes in the growth of sweet potato. Evidence of its positive contribution to growth was assayed successfully on IR 64 rice seedlings. Fungal endophyte isolates P3AL2c and P3BS1c have been discovered to improve growth of rice seedling without causing external manifestations of infection. This result has important implications to the agronomic management of both sweet potato and rice. However, further experiments must be made to make sure that these isolates are truly mutualistic symbionts and not latent pathogens. The successful application of these isolates and all other beneficial microorganisms as agronomic amendments depend on our understanding of their behaviour in the plant host and their interactions with other microorganisms in the soil. This understanding includes the study of the dynamics of fungal colonisation of plant tissues [18].
On a larger scale, studies regarding growth promoting endophytes of other crops may be profitable. Such fungal endophytes may be applied as potential bio-fertilizers to these crops with minimal environmental risks. Beneficial fungi with endophytic capabilities can be inoculated in roots or other plant tissues for the plant to enjoy the benefits these mutualists confer to their original plant hosts [18].The results of this research add to the growing effort to use screened fungal endophytes with growth promoting characteristics for possible inclusion among alternative modern technologies to support food production.
The study of morphological characteristics of a fungus provides valuable information for its identification. However, many fungi including fungal endophytes exhibit cryptic morphological characteristics necessitating the use of molecular tools and techniques [19]. DNA sequence analysis methods are objective, reproducible, and rapid means of identification, and thus gaining importance [20]. In this research, the phylogenetic affinities of the two growth promoting fungal endophyte isolates were explored using the 5.8s gene and flanking ITS genes. The generation of the phylogenetic tree in this research is crucial because BLASTn search alone cannot overcome the possibilities of statistical errors [7]. On the basis of sequence homology and phylogenetic results, P3AL2c fungal endophyte isolate is identified as Fusarium sp. and P3BS1c fungal endophyte isolate is identified as Emericella nidulans.
Acknowledgements
The author would like to thank Dr. Virginia C. Cuevas for her invaluable support in the conduct of this research and for funding. Dr. Genalyn Q. Diaz is also acknowledged for accommodating the researcher in her laboratory. The inputs Prof. Austria and Dr. Dizon in the preparation of this manuscript are also appreciated.
References
- Vega F.E., Posada F., Aime C., Pava-Ripoll M., Infante F.. and Rehner S. (2008) Entomopathogenic fungal endophytes. Biological control, 46: 72-82.
- Arnold A.E. (2007) Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biology Reviews, 21:51-66.
- Bacon C.W. (1993) Abiotic stress tolerances (moisture, nutrients) and photosynthesis in endophyte-infected tall fescue. Agriculture, Ecosystems and Environment, 44:123-141.
- Rodriguez R.J., Volkenburgh E.V., Hoy M., Wright L., Beckwith F., Kim Y. and Redman R.S. (2008) Stress tolerance in plants via habitat-adapted symbiosis. The ISME Journal, 2:404–416.
- Ownley B.H., Gwinn K.D. and Vega F.E. (2010) Endophytic fungal entomopathogens with activity against plant pathogens: Ecology and Evolution. BioControl, 55:113-128.
- Zabalgogeazcoa I. (2008) Fungal endophytes and their interaction with plant pathogens. Spanish Journal of Agricultural Research, 6 (Special issue): 138-146.
- Hamayun H., Khan S.A., Khan A.L., Ahmad A., Nawaz Y., Sher H. and Lee I.J. (2011) Gibberellin producing Neosartorya sp. CC8 reprograms chinese cabbage to higher growth. Scientia Horticulturae, 129:347-352.
- Götz, M., Nirenberg H., Krause S., Wolters H., Draeger S., Buchner A., Lottmann J. and Berg G. (2006) Fungal endophytes in potato roots studied by traditional isolation and cultivation-independent DNA-based methods. FEMS Microbiology Ecology, 58:404-413.
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., and Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25:3389-3402.
- Arnold A. and Lutzoni F. (2007) Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology, 88(3):541-549.
- Kimura M. (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16:111-120.
- Felsenstein J.(1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution, 39:783-791.
- Fuglie K.O. 2007. Priorities for sweet potato research in developing countries: results of a survey. Hortscience, 42(5):1200-1206.
- Lee E.H., Byun J.K. and Wilding S.J. (1985) A new gibberellin biosynthesis inhibitor, paclobutrazol (PP333), confers increased SO2 tolerance on snap bean plants. Environmental and Experimental Botany, 25(3):265-275.
- Khan A.L., Hamayun M., Kang S.M., Kim Y.H., Jung H.Y., Lee J.H., and Lee J.H. (2012) Endophytic fungal association via gibberellins and indole acetic acid can improve plant growth under abiotic stress: an example of Paecilomyces formosus LHL10. BMC Microbiology, 12:3 doi:10.1186/1471-2180-12-3.
- Hassan H.A.H. (2002) Gibberellin and auxin production plant root fungi and their biosynthesis under salinity-calcium interaction. Rostlinna vyroba, 48:101-106
- Khan S.A., Hamayun H., Yoon H., Kim H.Y., Suh S.J., Hwang S.K., Kim J.M., Lee I.J., Choo Y.S., Yoon U.H., Kong W.S., Lee B.M. and Kim J.G. (2008) Plant growth promotion and Penicillium citrinum. BMC Microbiology, 8:231 doi:10.1186/1471-2180-8-231
- Macia´ -Vicente J.G., Rosso L.C., A. Ciancio A., Jansson H.B. and Lopez-Llorca L.V. (2009) Colonisation of barley roots by endophytic Fusarium equiseti and Pochonia chlamydosporia: Effects on plant growth and disease. Annals of Applied Biology 155:391–401.
- Bhagobhaty R.K. and Joshi S.R. (2011) Multi-loci molecular characterisation of endophytic fungi isolated from five medicinal plants of Meghalaya, India. Mycobiology, 39(2):71-78.
- Hamayun M., Khan M., Khan A., Kim H., Chaudhary M., Hwang Y., Shinn D., Kim I., Kim S., Lee B. and Lee I. (2009) Gibberellin production and plant growth enhancement by newly isolated strain of Scolecobasidium tshawytschae. Journal of Microbiology and Biotechnology, 19(6):560–565.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences