miRPreditor: a Novel MiRNA Target Predictor Based on SVM with Feature Analysis
Zhisong He, Dijun Chen, Kuangyu Wang, Ming Chen
1Department of Bioinformatics, College of Life Sciences, Zhejiang University, Hangzhou 310058, China
2CAS-MPG Partner Institute for Computational Biology, SIBS, CAS, Shanghai 200031, China
3Statistical Genetics & Bioinformatics, North Carolina State University, Raleigh, NC 27695-7566, USA.
#These co-first authors contributed equally
- Corresponding Author:
- Tel: +86(0)571-88206612
Fax: +86(0)571-88206612
E-mail: mchen@zju.edu.cn
Abstract
MicroRNAs (miRNA) have been proven to serve as important post-transcription regulators in gene expression. To understand the function of miRNAs, it is necessary to figure out the target gene of miRNAs. Here we developed a novel miRNA target predictor, miRPredictor, which is based on support vector machine (SVM) combining with feature selection procedure. We considered different types of features including the flanking sequences of the potential targets and pattern information. The features selected were also analyzed to dig out the intrinsic mechanism of miRNA-target interaction. miRPredictor is available at http://bis.zju.edu.cn/ mirpredictor/.
Keywords
miRPredictor, miRNA target, SVM, Feature analysis.
1. Introduction
MicroRNAs are small non-coding RNAs with approximate length of 22nt. They bind to complementary region of mRNA to repress mRNA translation or mediate degradation of mRNA [1-3]. Thus, they serve as very important post-transcription regulators in gene expression, playing important roles in many cell processes such as development and cell division [4,5].
The choice between translation repression and mRNA destabilization is thought to depend on the degree of complementarity between miRNA and its target mRNA [6]. miRNAs behave differently between plants and animals. They tend to show nearly perfect complementarity to their targets in plants, while usually having mismatches, gaps or G:U wobble pairs in animals. Nevertheless, no matter in plants or animal, the complementarity and thermodynamic stability between miRNA and its target mRNA, especially the 5’ parts of miRNAs, are thought to be important in the recognition process.
To date, more than 8000 miRNAs in different species have been discovered and stored in the miRBase database [7-9], with new miRNAs still being found rapidly. Several experimental approaches, including miRNA microarray, miRNA target site mutation and miRNA gene silence with LNA (Locked Nucleic Acid), have been developed for study of relationships between miRNAs and their regulatory targets [10-14]. By now, thousands of those relationships have been demonstrated, some of which collected by the database miRecords [15]. However, the experimental identification processes are time-consuming and labor intensive. Given this difficulty, it is necessary to develop computational approach for accurate prediction of miRNA-target relationships.
Based on different rules of the binding between miRNAs and their corresponding target mRNA, computational methods have been developed and widely used in the miRNA studies [16]. One of the most typical types of methods, e.g. miRanda [17], TargetScan [18,19] and PicTar [20], predicts the binding sites of appointed miRNA mainly base on the complementarity between the 5’ terminal of miRNA and mRNA. Another type of methods, e.g. DIANA-microT [21] and RNAHybrid [22], uses a different strategy, seeking the most stable miRNA-mRNA heteroduplex structures. Some other approaches like Rna [22,23] use a pattern-based workflow which is different from others. Most of methods above also take evolutional conservation into consideration, focusing on the more conserved sites in mRNAs. This feature can be a double edged sword, reducing the rate of false positive while on the other hand limiting the results.
As described above, conservation analysis is involved in most miRNA target predictors. It indicates that many features and mechanisms of the interaction between miRNA and mRNA were not considered in the previous studies. In recent researches, besides the directly interacting sites in mRNA, their flanking sequences and some other characteristics are also related to the miRNA regulatory process, which are rarely considered in the previous methods [24,25].
Machine learning and data mining methods have been widely used in the computational biology and bioinformatics area. MiTarget is the first famous miRNA target prediction method based on machine learning approach [26]. It extracts features from miRNA-target pairs and classifies them to the regulatory or non-regulatory group by a trained SVM classifier. MirTarget2 [27] is another SVM-based miRNA-target predictor, considering different features from miTarget. Both the study declared acceptable results of prediction. It indicates that SVM is an effective tool in the miRNA target prediction.
Here, we present a novel method for miRNA target prediction based on SVM. Besides features in miTarget, additional features describing interactions between miRNAs and their target sites in mRNA are also involved. Feature selection was used for predictor optimization, and features in the optimized feature set were also analyzed. Our predictor, miRPredictor, finally obtains an overall correct rate of 85.81% in the 10-fold cross validation, which is better than both TargetScan and the predictor based on only features involved in miTarget
2. Materials and Methods
Dataset
In our study, data was extracted from the database miRecords15, which is a database of experimental identified interactions between miRNAs and mRNAs. The first version of both human and Drosophila melanogaster in miRecords were used, the total number of which is 121 and 1311, respectively. After excluding those redundant and incomplete examples, we eventually obtained 278 validated miRNA-target pairs, including 83 ones of D. melanogaster and 195 ones of human. They were used as positive samples in this study.
To gain the negative samples, data were extracted from literature listed by miRecords manually. MiRNA-target site pairs which were wetexperimental proved to be non-regulatory were firstly collected. To get more negative samples in order to improve specificity, we inferred more negative samples by considering interaction between miRNA and mRNA after site mutation. Consequently, we got a negative sample set with 194 examples, including 30 ones of D. melanogaster and 164 ones of human.
With all these samples, we constructed three datasets for our research: complete dataset with all samples we have, fruit fly dataset with the 113 samples from D. melanogaster, and human dataset with the 359 samples from human.
Support Vector Machine (SVM)
In our study, SVM [28,29] was used to classify a miRNA-target site candidate to a regulatory one or a non-regulatory one. SVM is one of the most popular machine learning approaches used in different fields including many biological researches. It constructs an optimized hyperplane in the feature space to maximize segregation between different types of samples, and predict a new sample’s type by mapping it to the feature space. To implement nonlinear classification, SVMs allow an implicit mapping of feature vectors into a high-dimensional, non-linear feature space, with the kernel function to calculate similarity between samples in acceptable time. This study used a radial basis function (RBF) as the kernel function:
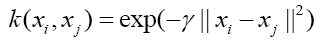
where xi, xj are the two feature vectors to be compare, and γ is the parameter determining the similarity level of features.
The SVM package LIBSVM [30] was used here to construct our SVM predictor. It was developed by Chang et al. and widely used in many areas. The package can be downloaded from https://www.csie.ntu.edu.tw/~cjlin/libsvm/.
Feature Vector Construction
For SVM classifier, each sample should be represented by a feature vector, which covers all aspect of the interaction between miRNAs and mRNAs. In this study, features can be categorized into 6 groups: structural features, thermodynamic features, position-based features, compositive features, secondary-structure features and patternbased features. The first three elements are also considered by miTarget [26], while the rest are novel ones imported for the prediction. Figure 1 shows the composition of all features involved.
Structural and thermodynamic features describe characteristics of the binding between miRNAs and their target sites. Structural features count the percentages of matches, mismatches, G:C matches, A:U matches, G:U wobble pairs and other mismatches from the five parts we considered, which consist of 5’ part (seed part), 3’ part of binding site alignment, the total alignment of binding site, the total alignment between miRNAs and 5’ flanking sequences of binding sites, and the total alignment between miRNAs and 3’ flanking sequences of binding sites. The former three parts of features are identical to what miTarget used, while the latter two are novel ones. Previous studies indicate the cooperation between miRNA binding sites adjacent to each other, and if any other possible binding site available in the flanking sequences, the tested binding site would be more likely to be a real one. Thermodynamic features are similar to structural ones, showing the free energy values of the five alignment structures as described above. Both structural and thermodynamic features are calculated by RNAduplex, one of the programs provided by Vienna RNA Package.
Position-based features are firstly introduced by miTarget, imitating the shape and mechanism of the seed pairing. It focuses on the matching situation at each specific position of miRNAs. Each position is represented by a vector with three dimensions, indicating A:U match, C:G match and G:U wobble pair, respectively. If this position is an A:U match, we could translate it into “1,0,0”, while C:G match, G:U wobble pair could be similarly coded into “0,1,0” and “0,0,1”, and “0,0,0” means mismatch. The first 20 nt of the appointed miRNA would be considered, so 60 features would be generated.
Previous studies show that the binding sites of miRNAs have some specific nucleotide composition [24,25], which cannot be clearly explained by known mechanisms. In other researches about nucleic acid, the nucleotide composition is also widely used [31]. In our study, we considered content of each nucleotide in the five parts of miRNA binding sites with the same way as the structural and thermodynamic features.
To regulate the target gene, the miRNA-binding site secondary structure is thought to play an important role, and should be thermodynamic stable enough [32]. Many classical miRNA target predictors such as RNAHybrid [22] come to their conclusions mainly based on the thermodynamic analysis of the miRNA-mRNA secondary structures. In our study, the candidate miRNA binding sites with their 100 nt length of flanking sequence on both sides are treated as a whole. Before miRNAs attached, he binding site together with its flanking components formed its own secondary structure, which was predicted by RNAcofold in Vienna RNA Package [33,34]. We counted the percentages of matches, mismatches, A:U matches, C:G mataches, G:U wobble pairs and other mismatches as parts of secondary structure features. We also calculated the free energy of the secondary structures before and after miRNAs’ binding by applying RNAcofold. The change of free energy in the binding process is also involved. Eventually, we obtained the 6+3=9 secondary structure features.
In several previous studies of miRNA target prediction, motifs are extracted from the sequences and considered as a series of important features [27,35]. Most of them count “words” in the binding site. It is simple, but these “words” rarely contain significant biological meanings. Rna22 [23] is miRNA target predictor based on motif discovery. It used Teiresias Algorithm to discover variable-length motif in known miRNAs. In this study, we adopt the same method to get results. Using the web server of Teiresias Algorithm at https://cbcsrv.watson.ibm.com/ Tspd.html, we obtained 228941 motifs. These motifs comprise a minimum length of L=4, have at least 30% of their positions specified (W=12) and appear a minimum of K=2 times in the input. Because mRNAs are reverse complement to miRNAs, the motifs should also be reversed and complemented to generate target site motifs. Here we consider four parts of miRNA binding sites, including the direct binding sites, 5’ flanking sequences of the binding sites, 3’ flanking sequences of the binding sites, and binding sites together with its flanking sequences. For each part, the valid pattern value which would be defined later is calculated. Firstly, motifs which exist in the appointed miRNA are selected as valid patterns. The number of these patterns would be counted and added together. Secondly, target site motifs corresponding to the valid patterns in the miRNA binding site would also be counted. Suppose there are n valid patterns in miRNA, N corresponding target site motifs in target site, the valid pattern value of this part of target site can be obtained by n/N.
Classifier Performance Evaluation
Cross-validation test and independent dataset test are widely in different fields for testing prediction quality in statistical prediction. In our research, three different categories of predictors were constructed with the same feature set but different training set. All predictors have gone through 10-fold crossvalidation. For the ones based on the fruit fly dataset or human dataset, besides 10-fold cross-validation test, an independent dataset test using samples of the other species was also utilized.
The results of tests can be described in different methods. Receiver operating characteristic (ROC) analysis, a plot of the true positive rate false positive rate, which is one of the most effective tools for evaluation, is also used to shows specificitysensitivity trade-off. Overall accurate rate is also calculated for comparison.
Feature Selection
Features with little distinction between different types of samples have negative effect to the performance of predictors 36. To improve the prediction results, feature selection should be used to obtain the optimal feature set. Moreover, by analyzing features in the optimal feature set, it is possible for us to interpret mechanism of miRNAmRNA binding better.
In this study, eight feature evaluation algorithms provided by Weka3 [36] are used to rank each feature in the complete feature set based on the complete dataset: Chi-Square Attribute Evaluation, Filtered Attribute Evaluation, Gain Ratio Attribute Evaluation, Information Gain Attribute Evaluation, OneR Attribute Evaluation, RelieF Attribute Evaluation, SVM Attribute Evaluation, Symmetrical Uncertainty (SU) Attribute Evaluation. Each feature would get a rank in every evaluation, when a smaller rank carries more importance. Total rank is defined as the sum of all 8 ranks. The evaluation scheme of the total rank is the same as single ranks.
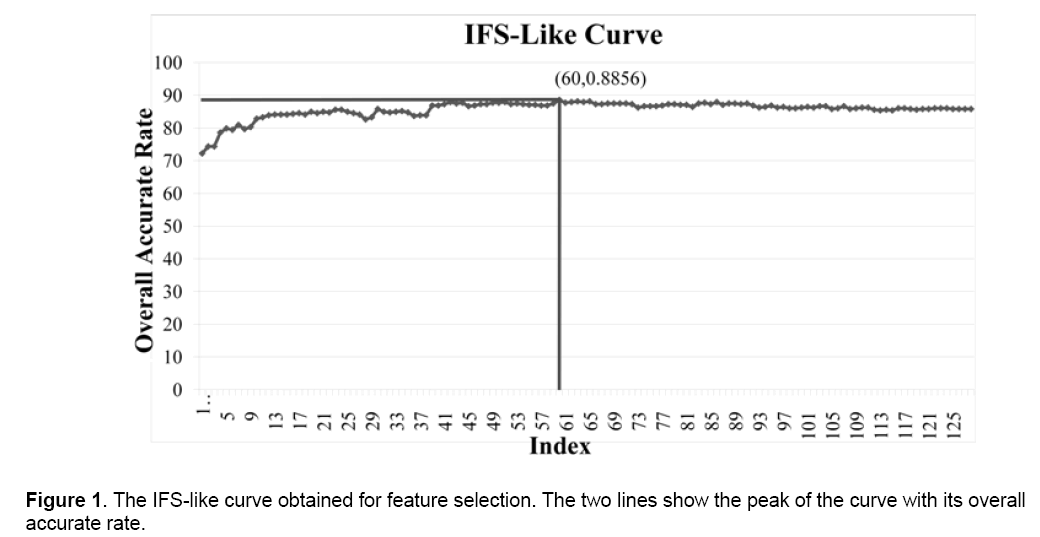
With the ranked list of all features, the next step is to determine the size of the optimal feature set. It is a process similar to Incremental Feature Selection (IFS) [37,38]. By adding features with the order of the list one by one, we could obtain N feature set where N is the total number of features and here N=128, while the i-th feature set is:
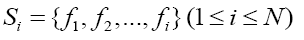
where fi is the i-th feature in the ordered feature list. Based on the complete dataset and the N feature set, N different SVM-based predictors were constructed. 10-fold cross-validation was used to test their performances. Soptimal = {f1,f2,…,fh}is regarded as the optimal feature set if the predictor based on it reaches the highest overall accurate rate in all predictors.
Statistical evaluation of features
To find out differences of features between positive and negative samples, Kolmogorov-Smirnov test (KS test) was used to test whether features in optimal feature set are different between positive and negative samples. K-S test is a form of minimum distance estimation used as a nonparametric test of equality of one-dimensional probability distributions used to compare a sample with a reference probability distribution (one-sample K–S test), or to compare two samples (two-sample K–S test). The null distribution of this statistic is calculated under the null hypothesis that the samples are drawn from the same distribution (in the two-sample case) or that the sample is drawn from the reference distribution (in the one-sample case). Here twosample case of K-S test was used. In this case, the K-S statistic is:
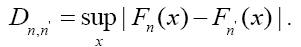
The null hypothesis is rejected at level α if:
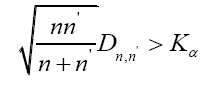
The null hypothesis is rejected at level α if:
3. Results and Discussions
Results of feature selection
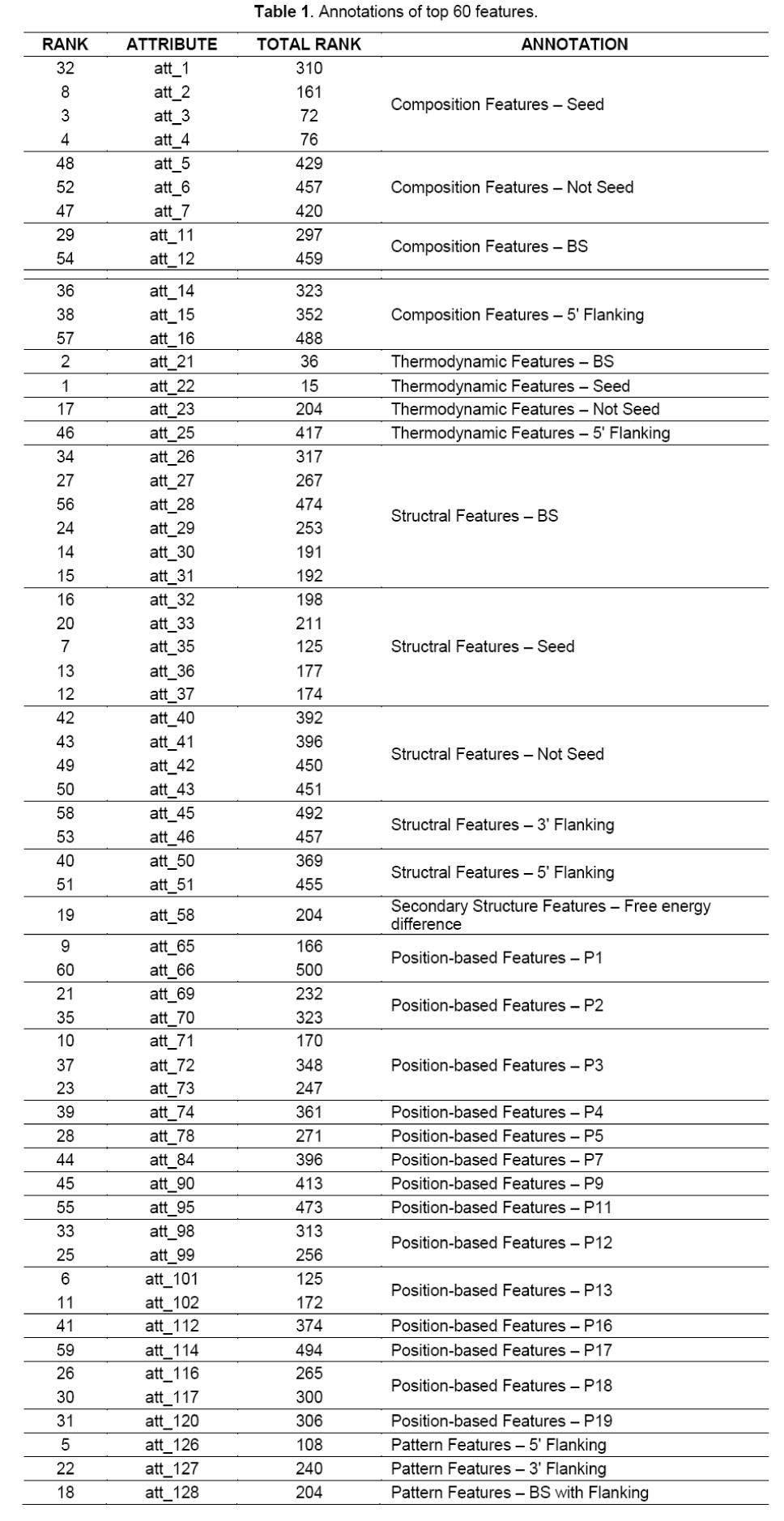
By integrating the eight feature evaluation algorithms in Weka3, we generated the feature rank list as supplemental material 1 shows. With this list, the IFS-like process was done and a curve could be drawn which the number of features as x-axis and the overall accurate rate of 10-fold cross-validation as y-axis (Figure 1). It is easy to find out the peak of curve with x-axis of 60, indicating the optimal feature set consists of the top 60 features in the ordered feature list. These features with annotation can be seen in Table 1.
Results of statistical evaluation of features
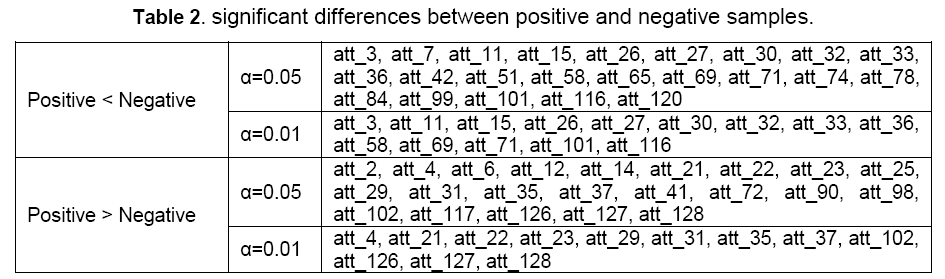
In our study, two different levels α was used: 0.01 and 0.05. At the level α=0.05, 45 in total 60 features in the optimal feature set show significant differences between positive and negative samples, while at the level α=0.01, the number becomes 26 (Table 2). At the level α=0.05, there are 23 features are larger in positive samples than that in negative ones, while the other 22 are just opposite.
Performances of the SVM predictors
After feature selection, we obtained the optimal feature set from the complete feature set. To compare with previous studies, features that also considered by miTarget are chosen to construct another feature set, miTarget feature set. Based on these 3 feature sets and the complete dataset, 3 different SVM predictors were constructed. 10-fold cross-validation was used for testing. The overall accurate rates are 85.81%, 85.17% and 88.56% for the predictors based on the complete feature set, miTarget feature set and optimal feature set, respectively.
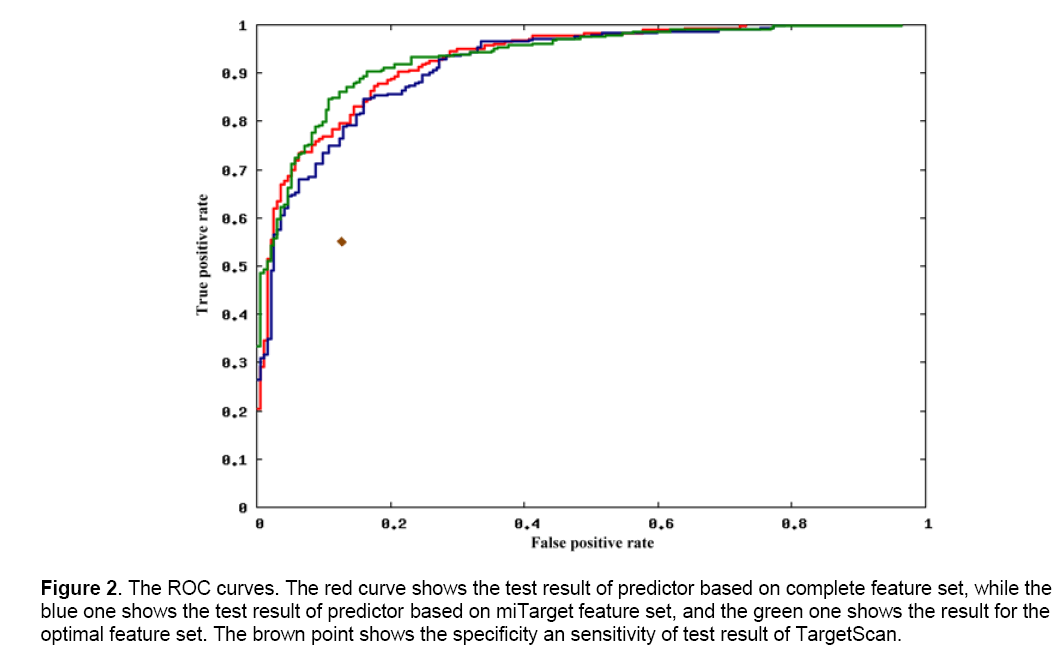
ROC curves were also used to analyze their performances (Figure 2). For the predictor with complete feature set (red), it gave an AUC (area under curve) of 0.9277. For the one with miTarget feature set (blue), its AUC is 0.9161, a little lower than the first one. In contrast, the predictor with optimal feature set gave out an AUC of 0.9318, which is higher than the other two. The three curves are close to each other in the area of high false positive rate (low sensitivity). However, in the low-false-positive-rate area (around 0.2), the predictor based on the optimal feature set get a obviously higher true positive rate than the other two. It indicates that the new features we introduced and the feature selection process are effective for improvement of predictor’s sensitivity and specificity.
Figure 2: The ROC curves. The red curve shows the test result of predictor based on complete feature set, while the blue one shows the test result of predictor based on miTarget feature set, and the green one shows the result for the optimal feature set. The brown point shows the specificity an sensitivity of test result of TargetScan.
Comparison between cross-validation test and cross-species independent dataset test
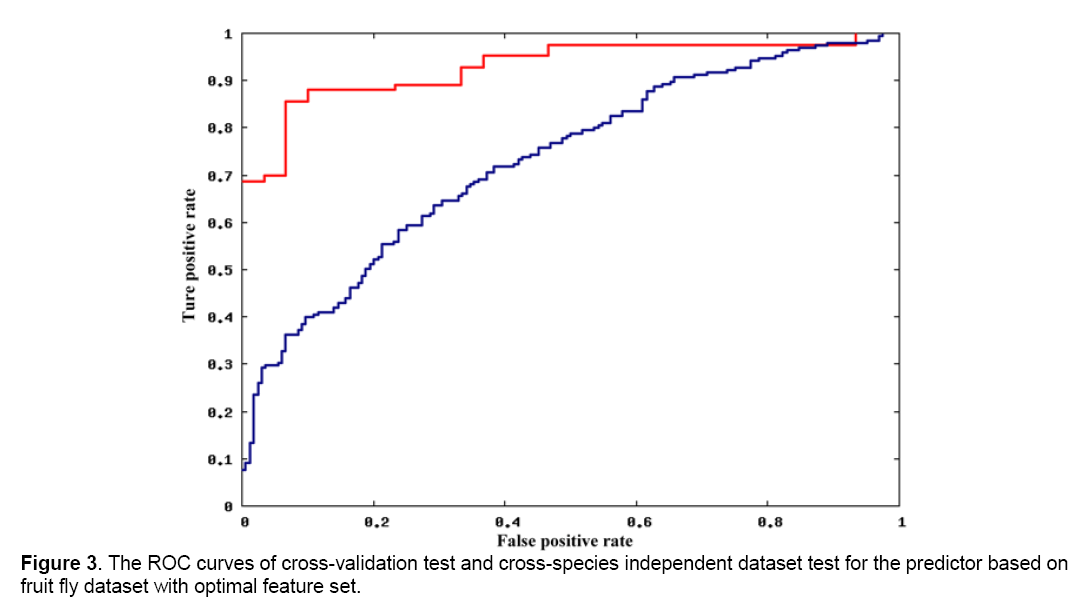
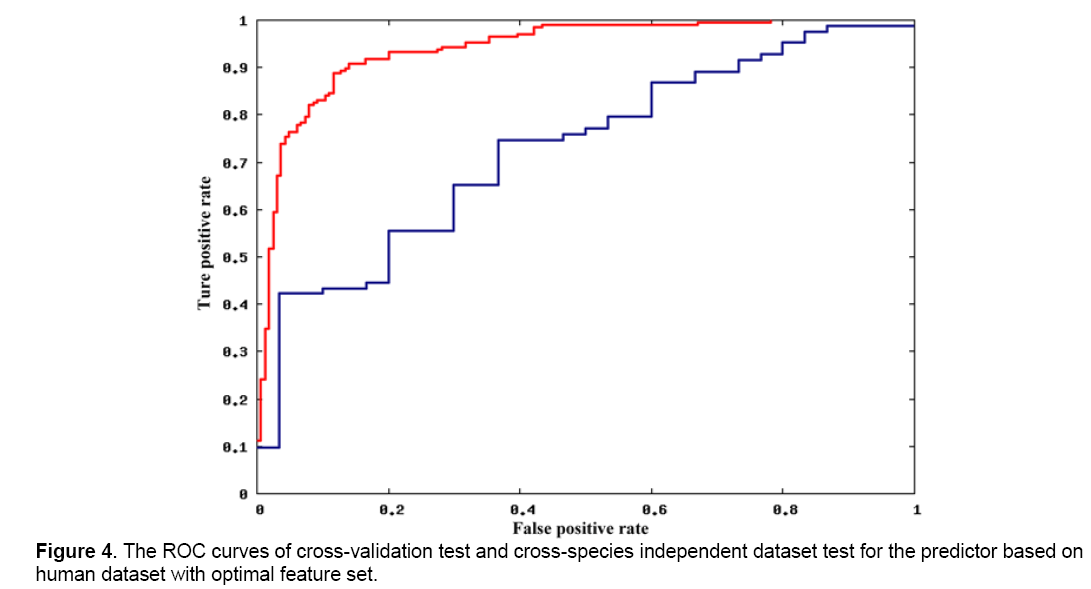
To study the different characteristics of miRNA-mRNA interactions between different species, we constructed two predictors based on the optimal features set and the two species-specific dataset. Both 10-fold cross-validation tests and independent dataset test with the other species’ dataset were processed and then compared. Figure 3 and Figure 4 show the ROC curves of testing with the training set of fruit fly dataset and human dataset, respectively. Obviously, for both predictors, 10-fold cross-validation test obtained a much better results. In fact, predictors based on the other two feature sets gave out an even larger difference. It indicates that there should be a large difference between fruit fly and human for features in optimal feature set, showing the large difference of miRNA-target recognition mechanisms between different species.
Comparison with TargetScan
There are several miRNA target predictor developed before. Lewis et al. developed TargetScan [18,19] firstly for mammalian miRNA target, which have several versions for different species such as TargetScanHuman for human and TargetScanFly for fruit fly now. It depends on the seed complementary mechanism and conservation among different species. It has been widely used and accepted by experimental validation. To evaluate our predictor more impersonally, after excluding the samples generated from mutation and the ones do not exist in TargetScan database, all the remained samples were tested by TargetScanHuman and TargetScanFly according to the species, and compared to the results of our predictors. For the total 402 samples with 272 positive and 130 negative ones, TargetScan correctly predicted 261 ones (Table 2), indicating the overall accurate rate of 64.93%, which is lower than the 10-fold cross-validation test of our predictor with the rate of 88.56%. The true positive rate of TargetScan is 0.5478, while the false positive rate is 0.1385, which is denoted by the brown diamond in Figure 2. When with the same specificity, our predictor could reach sensitivity of more than 0.75, which is better than TargetScan.
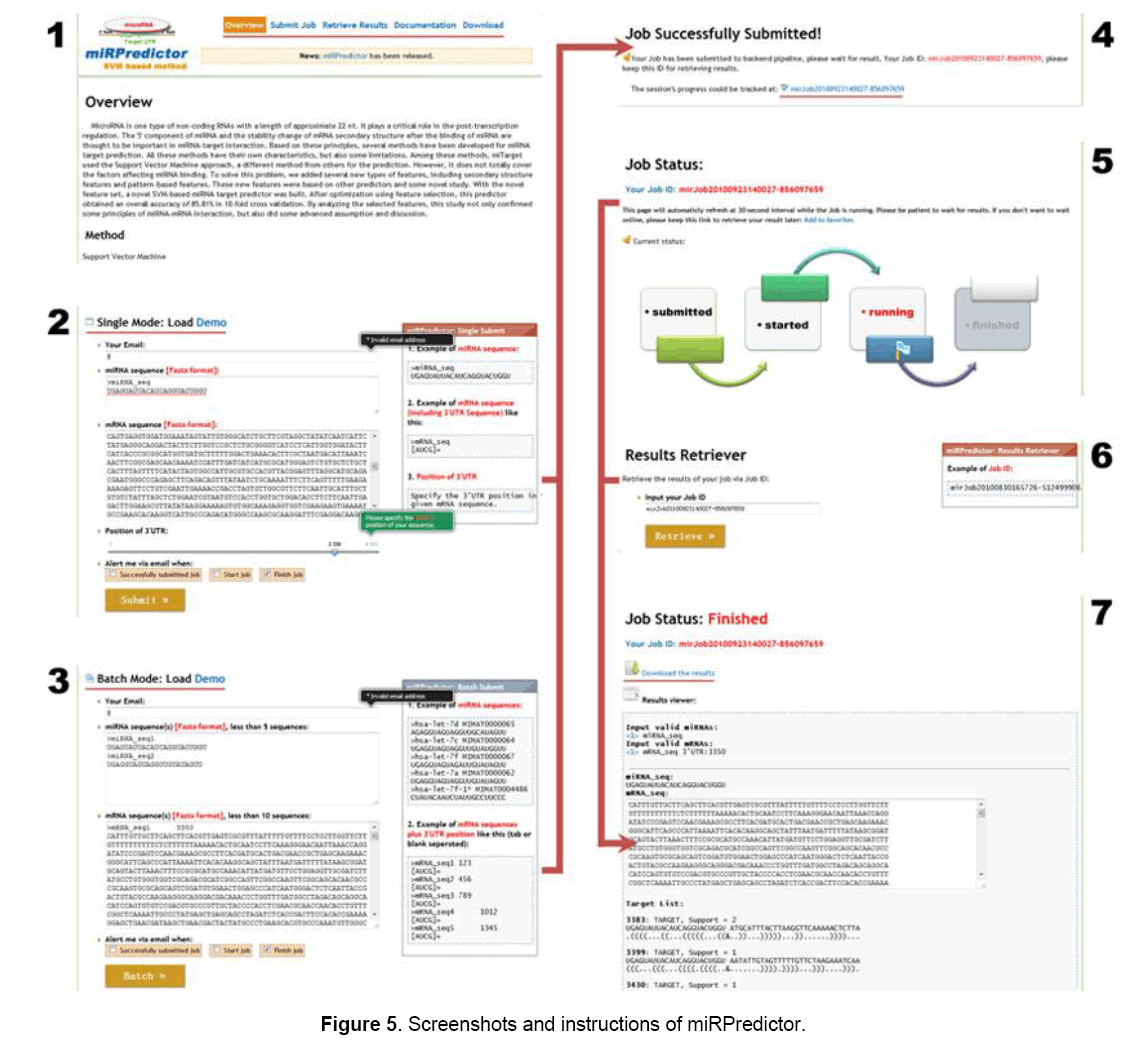
miRPredictor Implementation
miRPredictor’s Web service has been implemented on internet https://bis.zju.edu.cn/mirpredictor/ (Figure 5), free accessible to users. Its program can also be available for download.
Discussion
In our study, features were used to represent the miRNA-target interacting pairs, and different features indicate different aspects of mechanisms of miRNA-mRNA interaction. To learn more about this mechanism, we analyzed the features based on the ordered feature list output by feature evaluation process and the optimal feature set selected by feature selection process.
The top two features in the list are both thermodynamic features. The most important feature is the free energy of the secondary structure formed by seed region of miRNA and the target site, while the second one is the free energy of the structure formed by the whole miRNA and the target site. It indicates that thermodynamic stability of the miRNA-target heteroduplex structure, especially the 5’ part of miRNA, plays an important role in miRNA target recognition process. It is consistent with mechanisms provided by previous studies, and it is considered by a substantial part of miRNA target predictors which are based on thermodynamic analysis and seed region complementarity. In addition, the change of free energy of the mRNA secondary structure with the flanking sequences on each side after miRNA binding to is also selected to the optimal feature set with the rank of 17, which has been widely considered by previous predictors. In the K-S test, the free energy change of positive samples is significantly larger than that of negative ones (att_58 in Table 2) (its value is smaller in positive samples because it is a decrement). It also implies the effect of the flanking sequences of the direct target sites of miRNAs. These consistencies with previous studies support the basic view of miRNA-mRNA interacting mechanisms, that the thermodynamic characteristics play important roles in miRNA target recognition [6].
In the 60 features in the optimal feature set, 19 of them are structural features, indicating structural features to be an adapt type of features for miRNA target prediction. Most of the selected features are for the direct target site. However, 4 of them are for 5’ and 3’ flanking sequences of the target, and 1 of these 4 features (att_51 in Table 1 and Table 2, representing C:G match percentage in miRNAs with 5’ flanking sequences of target sites) shows a significant difference between positive and negative samples, indicating the cooperation of the nearby miRNA target candidates [24].
Position-based features were firstly introduced by miTarget26. They imitate the shape and mechanisms of pairing in different positions. As expected, for most of the positions in the seed region part of miRNA, at least one feature was selected. However, for some other parts of miRNAs, features were also selected, in particular the position 12 and 13 (att_98, att_99, att_101, att_102). These 4 features gained quite high ranks, and show significant difference between positive and negative samples in the K-S test. This result is consistent with what Grimson et al. discovered.
There are 13 composition features selected to the optimal feature set. Some of them even got high ranks such as att_2 (U content in miRNA seed region), att_3 (C content in miRNA seed region), att_4 (G content in miRNA seed region) in Table 1. It indicates a specific nucleotide compositional pattern exists in the miRNA target sites. Interestingly, the C content seems to be very important, which is selected in all the 4 clusters of composition features. In K-S test, all these 4 features of C content show significant differences between positive and negative samples, inferring some unknown mechanisms of the regulatory process. 3 of the 4 pattern-based features are selected.
Interestingly, quite many features of the 5’ flanking sequences of miRNA target site are selected to the optimal feature set, while in contrast, the number of features of the 3’ flanking sequences is much smaller. This result shows that, in the process of miRNA binding and regulating, the 5’ flanking sequences of the direct target is much more important than the 3’ flanking sequences. This asymmetry is somewhat similar to miRNA, where the 5’ part is more important than the 3’ part. This is a blank in the research of miRNA targeting. We inferred the key point to be the function of RISC protein. As previous studies show, miRNA combines with RISC to form miRNP complex to regulate expression of the target genes. The 5’ flanking sequence of miRNA target in mRNA may play an important role in the combining with miRNP and activating it to regulate the expression of the target gene post-transcriptionally.
Conclusion
In this paper, we introduce a novel SVM predictor, miRPreditor, for miRNA target prediction, and have shown its reliability in different aspects. Feature evaluation and selection is used to optimize the classifier and have gained a better result. The mechanisms of miRNA recognition and binding process are also discussed based on the results of feature selection and statistical analysis. Our results are consistent with previous studies, while on the other hand some new characteristics are discovered.
There are still some limitations, not only of our predictor, but also other computational methods for miRNA target prediction. The mechanism of miRNA function is not clear. The experimentally conformed data are deficient. Moreover, as our analysis indicates, the characteristics of miRNA function are species-specific. With more high-quality data covering more species, miRPredictor should gain a better result. Integrating results of different predictor together is also an effective way to improve accuracy of miRNA target prediction. accuracy of miRNA target prediction.
Acknowledgements
This work was supported by the National Natural Sciences Foundation of China [30771326, 30971743, 31050110121], the National High Technology Research and Development Program of China [2008AA10Z125], and the Program for New Century Excellent Talents in University of China [NCET-07-0740].
References
- Bartel D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116(2): 281-297.
- Filipowicz W., Jaskiewicz L., Kolb F.A., Pillai R.S. (2005) Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol, 15(3): 331-341.
- Sontheimer E.J., Carthew R.W. (2005) Silence from within: endogenous siRNAs and miRNAs. Cell, 122(1): 9-12.
- Ambros V. (2004) The functions of animal microRNAs. Nature, 431(7006): 350-355.
- Kong Y., Han J.H. (2005) MicroRNA: biological and computational perspective. Genomics Proteomics Bioinformatics, 3(2): 62-72.
- Bartel D.P. (2009) MicroRNAs: target recognition and regulatory functions. Cell, 136(2): 215-233.
- Griffiths-Jones S. (2004) The microRNA Registry. Nucleic Acids Res, 32 (Database issue): D109-D111.
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res, 34(Database issue): D140-D144.
- Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res, 36(Database issue): D154-D158.
- Friggi-Grelin F., Lavenant-Staccini L., Therond P. (2008) Control of antagonistic components of the hedgehog signaling pathway by microRNAs in Drosophila. Genetics, 179(1): 429-439.
- Lee D.Y., Deng Z., Wang C.H., Yang B.B. (2007) MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A, 104(51): 20350-20355.
- Li Y., Wang F., Lee J.A., Gao F.B. (2006) MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev, 20(20): 2793-2805.
- Ma L., Teruya-Feldstein J., Weinberg R.A. (2007) Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature, 449(7163): 682-688.
- Musiyenko A., Bitko V., Barik S. (2008) Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates prostein translation and invasiveness of prostate cancer LNCaP cells. J Mol Med, 86(3): 313-322.
- Xiao F., Zuo Z., Cai G., Kang S., Gao X., et al. (2009) miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res, 37(Database issue): D105-D110.
- Maziere P., Enright A.J. (2007) Prediction of microRNA targets. Drug Discov Today, 12(11-12): 452-458.
- Enright A.J., John B., Gaul U., Tuschl T., Sander C., et al. (2003) Marks, D. S., MicroRNA targets in Drosophila. Genome Biol, 5(1): R1.
- Lewis B.P., Burge C.B., Bartel D.P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120(1): 15-20.
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. (2003) Prediction of mammalian microRNA targets. Cell, 115(7): 787-798.
- Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., et al. (2005) Combinatorial microRNA target predictions. Nat Genet, 37(5): 495-500.
- Kiriakidou M., Nelson P.T., Kouranov A., Fitziev P., Bouyioukos C., et al. (2004) A combined computational-experimental approach predicts human microRNA targets. Genes Dev, 18(10): 1165-1178.
- Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. (2004) Fast and effective prediction of microRNA/target duplexes. RNA, 10(10): 1507-1517.
- Miranda K.C., Huynh T., Tay Y., Ang Y.S., Tam W.L., et al. (2006) A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell,126(6): 1203-1217.
- Didiano D., Hobert O. (2008) Molecular architecture of a miRNA-regulated 3' UTR. RNA, 14(7): 1297-1317.
- Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., et al. (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell, 27(1): 91-105.
- Kim S.K., Nam J.W., Rhee J.K., Lee W.J., Zhang B.T. (2006) miTarget: microRNA target gene prediction using a support vector machine. BMC Bioinformatics, 7: 411.
- Wang X., El Naqa I.M. (2008) Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics, 24(3): 325-332.
- Boser B.E., Guyon I.M., Vapnik V. (1992) In A training algorithm for optimal margin classifiers The fifth annual workshop on Computational learning theory Pittsburgh, Pennsylvania, United States.
- Vladimir N. (1998) Statistical Learning Theory Wiley.
- Chang C.C., Lin C.J. (2001) LIBSVM: a library for support vector machines.
- Yang Y., Wang Y.P., Li K.B. (2008) MiRTif: a support vector machine-based microRNA target interaction filter. BMC Bioinformatics, 9(Suppl 12): S4.
- Hofacker I.L. (2007) How microRNAs choose their targets. Nat Genet, 39(10): 1191-1192.
- Bernhart S.H., Tafer H., Muckstein U., Flamm C., Stadler P.F., et al. (2006) Partition function and base pairing probabilities of RNA heterodimers. Algorithms Mol Biol, 1(1): 3.
- Hofacker I.L., Fontana W., Stadler P.F., Bonhoeffer L.S., Tacker M., et al. (1994) Fast folding and comparison of RNA secondary structures. Monatshefte für Chemie, 125(2): 167-188.
- Wang X. (2008) miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA,14(6): 1012-1017.
- Witten I.H., Frank E. (2005) Data Mining: Practical machine learning tools and techniques. 2nd Edition ed.; Morgan Kaufmann: San Francisco
- Cai Y.D., Lu L. (2008) Predicting N-terminal acetylation based on feature selection method. Biochem Biophys Res Commun, 372(4): 862-865.
- Niu B., Lu L., Liu L., Gu T.H., Feng K.Y., et al. (2009) HIV-1 protease cleavage site prediction based on amino acid property. J Comput Chem, 30(1): 33-39.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences