Mapping of the Bovine Y Chromosome
Wan-Sheng Liu, F. Abel Ponce de León
Wan-Sheng Liu1,* and F. Abel Ponce de León2
1Department of Animal Biotechnology, College of Agriculture, Biotechnology and Natural Resources, University of Nevada, Reno, NV 89557, USA
2Department of Animal Science, College of Food, Agricultural, and Natural Resources Sciences, University of Minnesota, St. Paul, MN 55108, USA
Abstract
The bovine Y chromosome (BTAY) map has been developed considerably in recent years. There are approximately 260 DNA markers, including ~50 MS, 10 genes/ESTs, and ~200 BES, on BTAY. These markers together with their associated RH map and BAC fingerprinted contigs provide the basic materials for the future sequencing of BTAY. Several bovine homologs of human Y chromosome genes have been isolated. These genes are found to be expressed mainly or only in the testis, which opened the door for molecular approaches to the study of spermatogenesis and subfertility/infertility in bulls. The outcomes of such studies will probably lead to the design of new marker-assistant selection (MAS) strategies for male fertility to select sires at an early age in breeding programs.
Keywords
Y chromosome; mapping; bovine
1. Introduction
Sex determination in mammalian species is accomplished by the XY sex chromosome (Chr) mechanism. The Y Chr, carried by males only, plays an essential role not only in male sex development, but also in spermatogenesis and male fertility [1]. A small region located in the distal part of the short arm (p) of the Y Chr is known as the pseudoautosomal region (PAR) and the rest of the short and long arm (q) contain Y Chr male-specific sequences (MSY). These two regions have contrasting genetic properties. The X and Y Chrs. pair and recombine at the PAR during the pachytene stage of the first male meiotic division; the Y-specific and X-specific regions do not. The MSY region, comprising 95% of the DNA content of the Y Chr, can be further divided into two regions, euchromatic and heterochromatic. According to the human Y Chr sequence, the euchromatic region contains at least four different types of sequences [X-transposed (99% of similarity to the Xq21), X-degenerate (60-96% to the X), ampliconic, and centromere repetitive sequences] [2], and harbors all genes of the MSY, whereas the heterochromatic region contains Y-specific repetitive sequences. The absence of recombination with the X Chr in the MSY region at meiosis, the abundance of Y-specific repetitive sequences, the tendency of its genes to degenerate during evolution, and the functional coherence of its gene content in male growth, spermatogenesis and fertility [1] are some of the characteristics that make the Y Chr unique among all other nuclear Chrs. The absence of recombination makes genetic mapping of the MSY virtually impossible, and the depth, breadth and complexity of the repetitive sequences make sequencing extremely difficult. Therefore, mapping and sequencing strategies applied successfully elsewhere in the genome have faltered in the MSY [3], making the mammalian Y Chr a difficult target for linkage mapping and ultimately, sequencing. These difficulties led animal genome sequencing projects, including the Bovine Genome Sequencing Project (BGSP), to choose to sequence DNA from female animals [4-6]. To date, only the human, chimpanzee and mouse Y Chrs have been sequenced [1, 2, 7]. Our objectives for the past few years have been to characterize the organization of the bovine Y Chr, develop a bacterial chromosome tiling path to facilitate its sequencing and characterize bovine male-specific genes. Here we describe the progress achieved towards each objective.
2. The development of a BTAY-specific DNA library
Structurally, BTAY is the smallest Chr of the genome, comprising ~1.7% of the haploid genome. The estimated size of the entire Y Chr is ~51 Mb (3000 x 1.7%), and the euchromatic region is about half of the Y Chr. When the BTAY mapping project started ten years ago, there was little information available about the bovine Y Chr. In order to rapidly develop molecular genetic markers for BTAY, we have adapted a strategy of chromosomal microdissection and microcloning proposed by Saunders et al. [8] to dissect the whole Y Chr from bovine metaphase spreads [9]. The dissected Y Chr DNA fragments were purified, ligated with an adaptor, and then amplified by PCR with a primer that was complementary to the sequence in the adaptor [9]. A fraction of the amplified Y Chr inserts was used to generate the BTAY-specific DNA library, and another fraction was used for chromosome painting probes. The application of the Y Chr paints on bovine metaphase spreads by fluorescent in situ hybridization (FISH) allowed us to identify and localize the PAR on both BTAX and BTAY. The latter is localized in the telomeric region of the Yp, and of the Xq [10], rectifying the previous incorrect mapping of the PAR to BTAXp [11].
3. The development of BTAY microsatellite markers and the construction of a first-generation BTAY radiation hybrid map
Screening the BTAY-specific library with isotope-labeled (CA)12 oligos resulted in a total of 284 positive clones. After eliminating clones containing non-microsatellite Y Chr repetitive sequences, 118 clones were selected for sequencing, which identified 45 microsatellites (MSs) for BTAY [12]. Characterization of these MSs by PCR amplification of male and female bovine genomic DNA indicated that six mapped to the PAR, and 28 to the MSY region. The remaining 11 MSs amplified nonspecific bands and were difficult to map and thus, excluded [12]. Therefore, a total of 34 new MS markers were developed from the BTAY-specific library [12] (Table 1).
Besides our efforts to develop MS markers from the BTAY-specific library, several research groups worldwide [13-19] have also generated genetic markers for the Y Chr. To date, an additional 40 loci on BTAY are listed in the INRA BOVMAP Database (https://dga.jouy.inra.fr/cgi-bin/lgbc/loci_part.operl?MAPYN=Mapping&BASE=cattle&PARTIE=BTAY). Among these markers, 33 are DNA fragments [MS or sequence tagged site (STS)], and the remaining 7 markers are structural genes (Table 1).
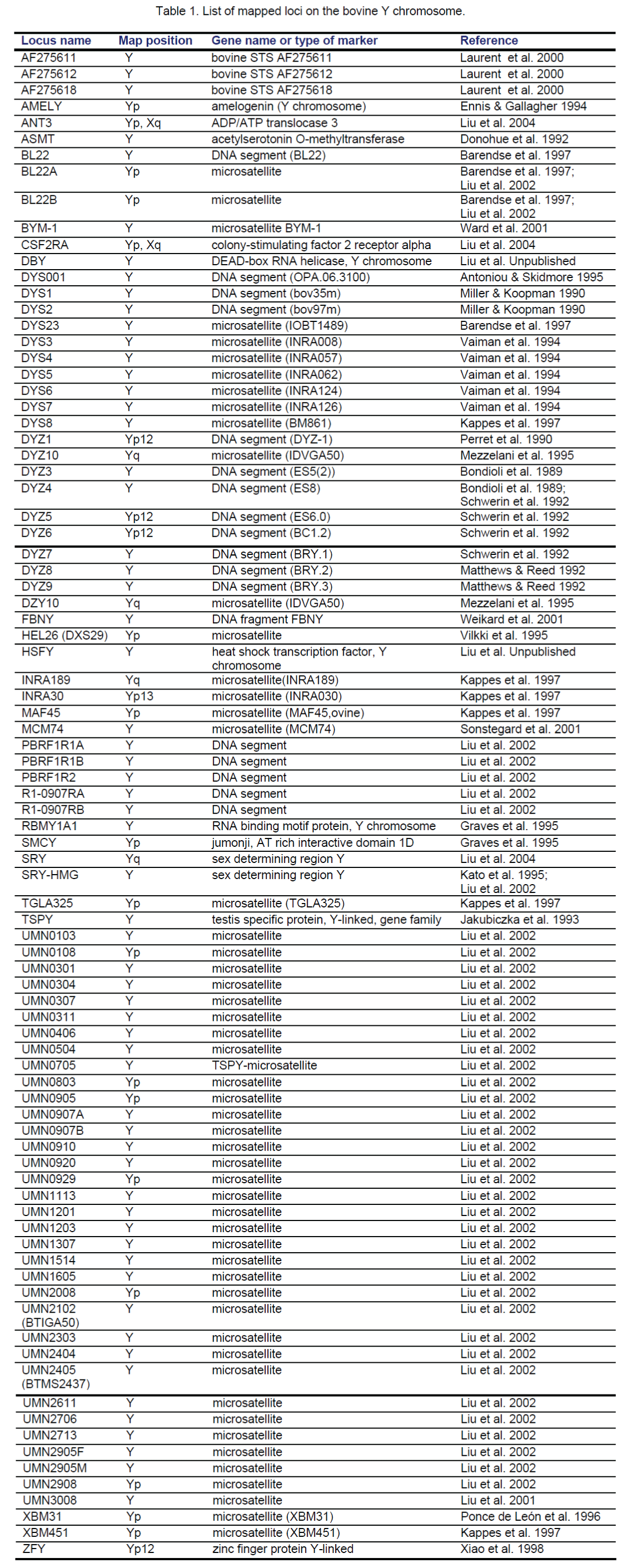
The absence of recombination at meiosis in MSY makes radiation hybrid (RH) mapping one of the best approaches to map this region. There are two BTAY RH maps reported so far. One is from our group with 33 markers on the map [12] using a 7000-rad cattle-hamster whole genome (WG) RH (SUNbRH7000-rad) panel [20]. The other assigned 11 markers using a 5000-rad cattle-hamster WG-RH (Illinois–Texas RH5000-rad) panel [21]. In the RH7000-rad map, a total of 62 markers, including 3 genes, 49 MS and 10 STSs were typed. Retention frequency (RF) of individual markers ranged from 18.5% to 76.5% with an average of 48.4%, in comparison to an average of 17.5% (3.3% to 53.3%) for non-Y Chr markers across the entire genome in the SUNbRH7000-rad panel [22]. It is worthwhile to note that over 40% of the typed Y markers have a RF of >55%, which were found to exist in multiple copies on the Y Chr and could not be mapped by the RH mapping method. At LOD score of 6, 13 markers were placed in the PAR with the AMELY gene proximal to the pseudoautosomal boundary (PB) and 20 markers, including SRY and TSPY genes, on the MSY of the RH7000-rad map [12,23].
4. Polymorphic Y Chr markers and Y haplotype
In 1997, Hanotte et al. [24] reported a polymorphic MS marker, INRA124, on BTAY. Later, Edwards et al. investigated four Y-specific MS (INRA124, INRA126, INRA189, and BM861) in different bovid species including domestic cattle, bison, mithan, swamp buffalo and yak, and found that these markers were highly polymorphic in the species studied [25]. We have assessed 38 Y Chr MS for polymorphisms in 17 unrelated bulls and one cow from six breeds (or crossbreeds) of domestic cattle [26]. Fourteen out of 38 MS were found to be polymorphic, and the remaining 24 were uninformative among the animals tested. Only one (INRA189) of the four MS that had shown polymorphisms in the study of Edwards et al. [25], was found to be polymorphic in our test samples. The reason for the relatively low rate of polymorphism in our analysis is most likely because the animals used in our study were from closely related cattle breeds (crosses). If such assessment is carried out in a large population or in more diversified cattle breeds, or in other bovid species, we predict that the 24 uninformative markers may prove to be polymorphic. Among the 14 polymorphic MS, the five PAR MS, on average, were more polymorphic (35.3%) than the nine MS on MSY (19.6%). These results are expected because the PARs on BTAY and BTAX recombine during meiosis, and therefore, the region is prone to generate more polymorphisms [12]. As discussed above, many of the MSY MS were multi-copies, whereas non-MSY MS were usually single copy. During genotyping, the multi-copy MS produces either ladder-like bands or a smear, preventing the identification and count of loci number. This is one of the unique features we found when doing Y Chr marker polymorphism analysis. The question is, are these multi-copy MS useful in identifying individual Y Chrs? Our experience with the sample of 17 bulls indicated that the ladder-like band (or smear) pattern was unique to individuals and/or breeds. Hence, the multi-copy MS were useful in Y Chr polymorphic analysis [26].
As the Y Chr is paternally inherited, it has been increasingly used to study the evolution and migration patterns of modern humans by using Y Chr haplotypes that have also been widely used in human forensic studies and paternity testing [27]. Our analysis of the 14 BTAY polymorphic MS indicated that each bull possessed a unique Y haplotype that could be used to identify itself or breed/crossbreed [26]. We believe that along with development of more polymorphic Y Chr markers, BTAY haplotypes will provide a powerful tool for paternity testing, analyzing diversity, distribution and lineage of the Y Chr, and studying the origin of domestic cattle as well as bovid species.
5. Genes on the Y Chr
For many years, it was assumed that the Y Chr was a wasteland carrying no genetic information apart from the sex-determining gene SRY. Recent work in the sequencing of the human Y Chr has revealed that there are 34 genes in the human PAR, and 156 known transcription units in the human MSY [2]. Comparative mapping among several species including human, mouse, cattle, sheep, dog, lemur and Sminthopsis macroura, has demonstrated that the Y Chr PAR genes are not conserved across mammalian species [28]. The variation of the PAR gene content and the origin and evolution of the PAR in mammals were explained by an “addition-attrition” theory, as proposed by Graves et al. [28-30]. Among the 156 transcription units on the human MSY, 78 are protein-coding genes that collectively encode 27 (18 single copy genes and 9 gene families) distinct proteins. The remaining 78 transcription units are non-coding transcripts [2]. These protein coding genes are classified into four groups. Group I contains one gene (SRY) that is involved in sex determination and is conserved among all mammals studied so far. Group II contains 15 genes (EIF1AY, CYorf15A and 15B, DBY, NLGN4Y, PCDH11Y, PRKY, USP9Y, RPS4Y1, RPS4Y2, SMCY, TBL1Y, TGIF2LY, TMSB4Y, ZFY and UTY) that are single copy, and expressed ubiquitously with a high degree of homology to genes present on the human X Chr. Four (USP9Y, DBY, UTY and TMSB4Y) genes from this group are clustered together and play a significant role in spermatogenesis and body size (stature) development [1]. The rest of the genes in this group have “housekeeping” functions. Group III contains genes (AMELY and GCY) that have been proposed to be related to the control of embryonic growth, stature and development of teeth [31,32]. Group IV contains nine genes (RBMY, DAZ, TSPY, CDY, BPY2, XKRY, PRY, HSFY and VCY). The most remarkable finding from the human Y Chr sequencing was the group IV gene families. They are all multi-copies, localized in the eight palindromes of the ampliconic sequences, expressed only in human testis, and are functionally coherent in spermatogenesis and fertility [1-3].
To date, a total of 14 orthologs (ANT3, CSF2RA, STS, AMELY, ASMT, RBM1A1, SMCY, ZFY, SRY, TSPY, DBY, HSFY, DAZ and CDY) of human Y Chr related genes have been analyzed to a certain degree in cattle (Table 1). Four (ANT3, CSF2RA, STS, AMELY) of these genes are localized in the PAR, the remaining genes in the MSY. These genes except for DAZ and CDY are physically mapped on the Y either by a comparative approach (SMCY, ASMT, RBM1A1, ZFY), or by restriction mapping and male-specific PCR (AMELY, TSPY), or by FISH and/or RH mapping (ANT3, CSF2RA, STS, SRY, DBY, HSFY and UTY) [12,23]. The DAZ and CDY gene families are not present on the BTAY, while their autosomal copies, DAZL, CDYL and CDYL2, do exist in the bovine genome (see below) [33].
To our knowledge, only three BTAY genes (AMELY, SRY, and TSPY) were previously cloned and characterized. The bovine AMEL genes reside on both the X- and Y Chrs and are expressed only in tooth buds. Alternative mRNA splicing generates at least seven messages, five from the AMELX primary transcript, and two from the AMELY [34,35]. The bovine SRY gene encodes a protein of 229 amino acids (aa), with sequence conservation between species, notably in the region of the high-mobility group (HMG) domain or HMG box. Outside of the HMG box, the bovine SRY structure shows greater resemblance to the human SRY than to the mouse Sry [36]. The bovine TSPY contains seven exons, and encodes a protein of 317 aa. Like the human TSPY, the bovine TSPY mRNA is a mixture derived from several seemingly functional genes organized in a tandem repeat array on BTAY, and is subject to differential splicing [18,37]. Both SRY and TSPY genes are expressed in bovine testis only. We are cloning the bovine orthologs of the human DBY, HSFY, UTY, BPY, PRY2, PRKY, USP9Y, VCY, RBMY, DAZ and CDY genes from a bovine testis cDNA library. These genes are all important for spermatogenesis and male fertility. Similar to the bovine AMEL gene, the bovine DEAD box protein gene also has a Y-copy (DBY) and an X-copy (DBX). The DBY encodes a protein of 660 aa, with a similarity of 88 and 89% to the human and orangutan DBY. According to the BGSP (build v2), the bovine DBX gene is predicted to transcribe at least seven transcription variants by alternative RNA splicing. The DBY protein is 85-89% similar to the different isoforms (1-7) of the DBX. The DBY is also expressed only in bovine testis. All other bovine Y Chr genes mentioned above are currently being sequenced and characterized.
In the human, the DAZ gene family has four copies (DAZ1-4) on the Y Chr and one copy (DAZ-like, or DAZL) on Chr 3 [38]. Deletions and single nucleotide polymorphisms (SNPs) identified in DAZ and/or DAZL have been linked to subfertility and infertility in several species including human [39], mouse [40], fly [41], and frog [42]. We have recently cloned the bovine DAZL gene, which is mapped to BTA1q by FISH [33]. We did not find any DAZ on BTAY, supporting a previous discovery that DAZL is present in all vertebrates, while DAZ is only in Old World monkeys and great apes [43, 44]. The bovine DAZL contains 11 exons, encodes a protein of 295 aa, and is highly (96%) conserved when compared to human and mouse DAZL. Two transcript variants were found for the bovine DAZL, which are expressed in bovine testis only, while the human and mouse DAZL are expressed in both male and female gonads [39,40]. We have recently identified 16 SNPs for the bovine DAZL gene. A preliminary association study indicated that these SNPs are associated with bull fertility. Similarly, there are four copies of the CDY gene on the human Y Chr, and two copies, CDYL (CDY-like) and CDYL2, on human autosomes [2,45]. It is believed that the progenitor of the gene family duplicated to generate CDYL and CDYL2, and CDY arose by retroposition of CDYL to the Y Chr and was retained only in simian mammals [45,46]. That explains why our work on bovine and another study in mice [46] found only the autosomal genes CDYL and CDYL2, but not CDY on the bovine or mouse Y Chrs. The bovine CDYL and CDYL2 are highly similar to the human orthologs at both mRNA (81 and 82%) and protein (89 and 94%) levels. However, the similarity between the bovine CDYL and CDYL2 proteins is low (41%). The bovine CDYL and CDYL2 genes were assigned by RH mapping to bovine Chr 23 and Chr 18, respectively [47]. Sequence analyses indicated that there are at least four transcript variants that yield three protein isoforms for the bovine CDYL gene. Expression analysis in different bovine tissues showed that genes in the CDYL family are abundantly expressed in testis [47]. A similar expression pattern was also observed for the human and mouse CDYL gene family [45, 46].
6. BTAY-specific BAC isolation and BAC fingerprinted contig map
To construct a high-resolution bacterial artificial chromosome (BAC) contig map for the BTAY euchromatic region, a male bovine BAC library (RPCI-42) was screened with probes of the microdissected bovine Y Chr DNA fragments as well as ~40 Y Chr gene PCR products generated either from the known bovine or from the human and mouse holandric genes. The microdissected bovine Y Chr DNA probe identified ~1300 positive clones. Altogether, these screenings resulted in the isolation of ~1800 Y Chr specific BAC clones. Based on the size of the Y Chr and an estimated 5.9x coverage of the Y Chr in the RPCI-42 BAC library (164 kb average/BAC clone), the number of isolated BAC clones mathematically covers the entire bovine Y Chr [48].
Isolated BAC clones were then fingerprinted by an agarose gel method [48]. Gel images and fingerprint patterns were analyzed using the IMAGE and FPC software (https://www.sanger.ac.uk/ Software/). A FPC database for BTAY was developed. A contig map built for the Y-specific BAC clones was completed at a tolerance of 7 with cutoff values of 1e-10 from which 37 contigs (range of clones in contigs from 3 to 925) and 750 singletons were observed. FISH of representative BAC clones from the contigs demonstrated that the largest contig containing 925 clones is situated on the proximal region of Yq [48].
7. BAC-end sequencing, STS mapping and the development of a second-generation BTAY RH map
Approximately 1350 BACs of BTAY have been sequenced either from one end (1150 clones) or both ends (200 clones). About 85% of the BAC-end sequences (BESs) are readable with an average of 500 bp clean sequence per read. Although initial BLAST searches against GenBank revealed that roughly one third of sequences were unique, a significant number were subsequently identified as novel BTAY-specific repetitive sequences. Approximately 500 STS PCR primers were designed from the putative unique BESs, and have been RH typed with the SUNbRH7000-rad panel [22]. We found that 43.6% STSs have a RF of less than 50%, and are therefore, initially considered putative unique sequences or lower copy markers on the Y Chr. As 33.6% STSs have a RF of more than 50%, we consider them as repetitive sequences or multiple copy (gene) sequences on the Y. The rest, 22.8% STSs, either did not amplify a product or showed weak products in the RH mapping. Construction of a second-generation BES-based BTAY RH7000-rad map is underway. Once this RH7000-rad map is finished, a minimal tiling path of BAC clones that cover the entire euchromatic region in MSY will be generated in combination with the BTAY FPC database, and used as the basic materials for Y Chr sequencing.
8. Conclusion
The Y Chr is unique by the absence of recombination, along 95% of its length, with X Chr in male meiosis, the accumulation of male reproduction-related genes, and the tendency of its genes to degenerate during evolution. The Y Chr has become a genetic junkyard, accumulating mutations and losing much of its genetic material [49]. Unlike the X Chr, whose gene content is highly conserved in mammalian species, Y Chr genes are poorly conserved.
The bovine Y Chr mapping is one of the most advanced Y Chr projects among all farm animal species, with a development of over 260 markers including ~50 MS, 10 genes/ESTs, and ~200 BES. At present, bull fertility has not been studied at the molecular level because of the lack of molecular genetic markers and diagnostic tools. The cloning of the bovine Y Chr genes has opened the door for molecular study of spermatogenesis and subfertility/infertility in bulls. This will significantly improve the design of new marker assistant selection (MAS) strategies using Y-related gene markers as an aid to select sires at an early age in a breeding program, and will eliminate potential genetic defects associated with reduced fertility. The possibility to construct highly informative Y Chr haplotypes will also have a significant impact on researches in paternity testing, breed formation, origin and evolution of domestic cattle and bovid species.
Acknowledgements
This work was supported by grants from USDA, CSREES to W.-S. Liu and F.A. Ponce de León (No. 2005-35205-15455), to F.A. Ponce de León and W.-S. Liu. (No.2002-35205-11627), and to F.A. Ponce de León (No 96-35205-3757).
References
- Lahn B.T., Page D.C. (1997) Functional coherence of the human Y-chromosome. Science, 278: 675-680.
- Skaletsky H., Kuroda-Kawaguchi T., Minx P.J., et al. (2003) The male-specific region of the human Y Chr is a mosaic of discrete sequence classes. Nature, 423(6942): 825-837.
- 3Tilford C.A., Kuroda-Kawaguchl T., Skaletsky H. et al. (2001) A physical map of the human Y-chromosome. Nature, 409: 943-945.
- 4Krzywinski M., Wallis J., Gosele C., et al. (2004) Integrated and sequence-ordered BAC- and YAC-based physical maps for the rat genome. Genome Res, 14(4): 766-779.
- 5Waterston R.H., Lindblad-Toh K., Birney E., et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature, 420(6915): 520-562.
- 6Lander E.S., Linton L.M., Birren B., et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409(6822): 860-921.
- 7Kuroki Y., Toyoda A., Noguchi H., et al. (2006) Comparative analysis of chimpanzee and human Y chromosomes unveils complex evolutionary pathway. Nat Genet, 38: 158-167.
- 8Saunders R.D., Glover D.M., Ashburner M., et al (1989) PCR amplification of DNA microdissected from a single polytene chromosome band: a comparison with conventional microcloning. Nucleic Acids Res, 17(22): 9027-9037.
- 9Ponce de León F. A. (1996) Microdissected chromosome libraries for livestock species. Arch Zootech, 45: 165-174.
- 10Ponce de León F.A., Carpio C. (1995). Identification of the bovine X chromosome pseudoautosomal region. 9th North American Colloquium on Domestic Animal Cytogenetics & Gene Mapping, Texas A&M University (Abstract) pp. 8.
- 11Barendse W., Armitage S.M., Kossarek L.M., et al. (1994) A genetic linkage map of the bovine genome. Nat Genet, 6(3): 227-35.
- 12Liu, W.S., Mariani, P., Beattie, C.W., et al. (2002) A radiation hybrid map for the bovine Y Chromosome. Mamm Genome, 13: 320-326.
- 13Bondioli K.R., Ellis S.B., Pryor J.H., et al. (1989) The use of male-specific chromosomal DNA fragments to determine the sex of bovine preimplantation embryos. Theriogenology, 31: 95-104.
- 14Miller J.R., Koopman M. (1990) Isolation and characterization of two male-specific DNA fragments from the bovine genome. Anim Genet, 21: 77-82.
- 15Matthews M.E., Reed K.C. (1991) A DNA sequence that is present in both sexes of Artiodactyla is repeated on the Y chromosome of cattle, sheep, and goats. Cytogenet Cell Genet, 56: 40-44.
- 16Vaiman D., Mercier D., Moazami-Goudarzi K., et al (1994). A set of 99 cattle microsatellites: characterization, synteny mapping, and polymorphism. Mamm Genome, 5: 288-297.
- 17Cui X., Kato Y., Sato S., et al. (1995) Mapping of bovine Sry gene on the distal tip of the long arm and murine Sry on the short arm of the Y Chr by the method of fluorescence in situ hybridization (FISH). Anim Sci Technol (Jpn), 66: 441-444.
- 18Vogel T., Borgmann S., Dechend F., et al. (1997) Conserved Y-chromosomal location of TSPY in Bovidae. Chromosome Res, 5(3): 182-185.
- 19Xiao C., Tsuchiya K., Sutou S. (1998) Cloning and mapping of bovine ZFX gene to the long arm of the X-Chr (Xq34) and homologous mapping of ZFY gene to the distal region of the short arm of the bovine (Yp13), ovine (Yp12-p13), and caprine (Yp12-p13) Y chromosome. Mamm Genome, 9(2): 125-130.
- 20Mariani P., Sugimoto Y., Beattie C.W. (2000) A new whole-genome radiation hybrid (WG-RH) panel for cattle. ISAG 2000, B082.
- 21Amarante M.R.V., Womack J.E. (2004) A preliminary radiation hybrid map of bovine Y chromosome. PAG-XII, P624.
- 22Itoh T., Watanabe T., Ihara N., et al. (2005) A comprehensive radiation hybrid map of the bovine genome comprising 5593 loci. Genomics, 85(4): 413-24.
- 23Liu W.S., Ponce de León F.A. (2004) Assignment of Sry, Ant3 and Csf2ra to the bovine Y Chr by FISH and RH mapping. Anim Biotechnology, 15: 103-109.
- 24Hanotte O., Okomo M., Verjee Y., et al. (1997) A polymorphic Y chromosome microsatellite locus in cattle. Anim Genet, 28(4): 318-319.
- 25Edwards C.J., Gaillard C., Bradley D.G., et al. (2000) Y-specific microsatellite polymorphisms in a range of bovid species. Anim Genet, 31: 127-130.
- 26Liu W.S., Beattie C.W., Ponce de León F.A. (2003) Bovine Y Chr microsatellite polymorphisms. Cytogenetics and Genome Research. 102: 53-58.
- 27Quintana-murci L., Krausz C., Mcelreavey K. (2001) The human Y chromosome : function, evolution and disease. Forensic Sci Int, 118: 169-181.
- 28Graves J.A.M., Wakefield M.J., Toder R. (1998) The origin and evolution of the pseudoautosomal regions of human sex chromosomes. Hum Mol Genet, 7(13): 1991-1996.
- 29Graves, JA. (1995) The origin and function of the mammalian Y Chr and Y-borne genes—an evolving understanding. BioEssays 17: 311-321.
- 30Graves, JA. (1998) Evolution of the mammalian Y Chr and sex-determining genes. Journal of Experimental Zoology, 281: 472-481.
- 31Fincham AG, Hu YY, Lau E, Pavlova Z, Slavkin HC, Snead ML.(1990) Isolation and partial characterization of a human amelogenin from a single fetal dentition using HPLC techniques. Calcif Tissue Int, 47(2): 105-11.
- 32Kirsch S, Weiss B, Zumbach K, Rappold G. (2004) Molecular and evolutionary analysis of the growth-controlling region on the human Y chromosome. Hum Genet, 114(2): 173-181.
- 33Liu W.S., Wang A., Uno Y., et al. (2007) Genomic structure and transcript variants of the bovine DAZL gene. Cytogenet Genome Res, 116 (1-2): 65-71.
- 34Gibson C., Golub E., Herold R., et al. (1991) Structure and expression of the bovine amelogenin gene. Biochemistry 30: 1075-1079.
- 35Yuan Z.A., Collier P.M., Rosenbloom J., et al. (1996) Analysis of amelogenin mRNA during bovine tooth development. Archs Oral Biol, 41: 205-213.
- 36Daneau I., Houde A., Ethier J.F., et al. (1995) Bovine SRY gene locus: cloning and testicular expression. Biol Reprod. 52(3): 591-599.
- 37Vogel T., Dechend F., Manz E., et al. (1997) Organization and expression of bovine TSPY. Mamm Genome, 8(7): 491-496.
- 38Cauffman G., Van de Velde H., Liebaers I., et al. (2005) DAZL expression in human oocytes, preimplantation embryos and embryonic stem cells. Mol Hum Reprod, 11: 405-411.
- 39Teng Y.N., Lin Y.M., Lin Y.H., et al. (2002) Association of a single-nucleotide polymorphism of the deleted-in-azoospermia-like gene with susceptibility to spermatogenic failure. J Clin Endocrinol Metab 87: 5258-5264.
- 40Ruggiu M., Speed R., Taggart M., et al. (1997) The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature 389: 73-77.
- 41Eberhart C.G., Maines J.Z., Wasserman S.A. (1996) Meiotic cell cycle requirement for a fly homologue of human Deleted in Azoospermia. Nature 381: 783-785.
- 42Houston D.W., King M.L. (2000) A critical role for Xdazl, a germ plasm-localized RNA, in the differentiation of primordial germ cells in Xenopus. Development 127: 447-456.
- 43Cooke H.J., Lee M., Kerr S., et al. (1996) A murine homologue of the human DAZ gene is autosomal and expressed only in male and female gonads. Hum Mol Genet 5: 513-516.
- 44Saxena R., Brown L.G., Hawkins T., et al. (1996) The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat Genet 14: 292-299.
- 45Lahn, B. T. and D. C. Page (1999). Retroposition of autosomal mRNA yielded testis-specific gene family on human Y chromosome. Nat Genet, 21(4): 429-33.
- 46Dorus, S., S. L. Gilbert, et al. (2003). The CDY-related gene family: coordinated evolution in copy number, expression profile and protein sequence. Hum Mol Genet, 12(14): 1643-50.
- 47Wang A.H., Ponce de Leon F. A., Liu W.S. (2007) The bovine CDYL gene family: cloning, mapping and expression analysis. PAG-XV, P530.
- 48Liu W.S., Ponce de León F.A. (2003) Development of a BAC-based physical map of the euchromatic region of the bovine Y-chromosome. PAG-XI. P586.
- 49The Y chromosome: goldmine and junkyard. News @Nature.com (https://www.nature.com/news/2001/ 010215/full/010215-5.html) Feb 12, 2001.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences
