Isopentenyl Phosphate Kinases are Ubiquitous and Copy Numbers are Conserved in Plant Genomes
Ben Hu, Heng Yao, Yulong Gao, Ran Wang, Feng Li, Zefeng Li, Lifeng Jin and Yanxiang
Ben Hu1,2,3#, Heng Yao4#, Yulong Gao4, Ran Wang3, Feng Li3, Zefeng Li3and Lifeng Jin3*
1School of Chemical Engineering, Dalian University of Technology, Dalian 116024, P.R. China
2Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, P.R.China
3Zhengzhou Tobacco Research Institute of CNTC, Zhengzhou 450001, P.R. China
4Yunnan Academy of Tobacco Agricultural Sciences, Kunming 650021, P.R. China
#Both authors contributed equally to this work.
*Corresponding Author:
Lifeng Jin Zhengzhou Tobacco Research Institute of CNTC Zhengzhou 450001, P.R. China Tel: 13592582785 E-mail: jin_lf@126.com
Received: December 23, 2019; Accepted: January 22, 2020; Published: January 29, 2020
Citation: Ben Hu, Heng Yao, Yulong Gao, et al. Isopentenyl Phosphate Kinases are Ubiquitous and Copy Numbers are Conserved in Plant Genomes. Electronic J Biol. 16:1
Abstract
Isopentenyl phosphate kinase (IPK) is a recently discovered enzyme playing a key role in mevalonate pathway in isoprenoid biosynthesis, while systematic investigation of IPKs in plants is lack Here, through genome-wide identification and analysis of IPKs, we showed that IPKs gene are ubiquitously present in plant genomes. All IPKs protein previously identified had AAK (Amino Acid Kinase) domain. From 35 plant species with genome assembly data available, we extracted all AAK family members. Using OrthoMCL, we identified a group of 37 sequences in which Arabidopsis IPK protein was included. Further analysis showed that each peptide sequence in this group has a His residue which is a signature of IPK enzyme, indicating that the genes in this group were IPKs protein. Not like these in other domains of life which showed spotty distribution over the tree of life, virtually all plant genomes we analyzed here had IPK genes. Further, copy numbers of IPKs gene were very conserved in that no higher than 2 copies remained in each plant genome. Plant IPKs formed a distinctive clade in phylogenetic tree of plant AAK gene family, and had a phylogenetic topology conformed to that of plant species. Our results indicate IPK plays important roles in plant physiology and is conserved in plant evolution. The IPKs gene we identified here would provide new molecular targets for characterization of plant mevalonate pathway, and shed light on biochemistry of plant isoprenoids biosynthesis.
Keywords
Isopentenyl phosphate kinase; Mevalonate pathway; Isoprenoids; Evolution; Genome
Abbreviation
HMM: Hidden Markov Model; IPK: Isopentenyl Phosphate Kinase; IPP: Isopentenyldiphosphate; MVA: Mevalonate; MEP: Methyl Erythritol; AAK: Amino Acid Kinase; OrthoMCL: Ortholog Markov Clustering Algorithm
1. Introduction
Isoprenoids constitute a large group of biologically active metabolites in living cells. Cells invariably require two five-carbon (C5) building blocks, isopentenyldiphosphate (IPP) and its isomer, dimethylallyldiphosphate (DMAPP), as precursors for isoprenoids. To biosynthesize these two compounds, two independent pathways, the mevalonate (MVA) pathway and the methylerythritol (MEP) pathway, are both present and functionally effective in plants, which are not like most other organisms where only one pathway or the other is utilized [1-3].
Regarding MVA pathway, Plants are also unique as an alternative route is present to this classical pathway. Following mevalonate 5-phosphate (MVAP) biosynthesis, the classical MVA pathway phosphorylates MVAP to produce diphosphomevalonate (MVAPP) by phosphomevalonate kinase (PMK), and then uses mevalonate-5-diphosphate decarboxylase (MDD) to decarboxylate MVAPP and produce IPP, C5 isoprenoid building block [4,5]. Genes encoding PMK or MDD were failed to be identified in archaea, which were intriguing, because isoprenoids constitute essential parts of archaea lipid membrane [6]. Identification of isopentenyl phosphate kinase (IPK) in Methanocaldococcusjannaschii led to a proposal of presence of an alternative MVA pathway in archaea [4,7]. In this pathway, firstly, MVAP was decarboxylated by a yet unidentified enzyme; next, phosphorylation led to production of IPP catalyzed by IPK protein [2-8]. IPK gene was originally thought to be restricted in archaea as PMK and MDD were only missing in archaea where other components of MVA pathway were present, and virtually no IPK gene was identified by approaches based on homology [4,9]. By using more sensitive searching approaches, Dellas et al. identified homologs of IPK proteins in bacteria and eukarya in addition to archaea, which were confirmed to be enzymatically functional [5,10]. Interestingly, despite a spotty phylogenetic distribution of homologs in fungi and animals, IPK sequences were identified in all 15 plants surveyed by Dellas et al. [10].
Here, through using a comprehensive identification of AAK family (Amino Acid Kinase) members, followed by orthoMCL(Ortholog Markov Clustering Algorithm) grouping, we identified 37 putative IPK sequences from 35 plants with genome assembly data available. Through sequence and phylogenetic analysis, we discussed their evolutionary and physiological implications.
2. Materials and Methods
2.1 Identification of AAK family members
37 plants with genome sequences available publicly were chosen for IPK sequence mining, which represent key stages in plant evolution history (Figure 1). The 15 plant IPKs identified by Dellas et al. and Arabidopsis were used as queries for BLAST searches, which were performed in a standalone mode using BLASTp included in BLAST+ tools (v2.2.30) (http://www.ncbi.nlm.nih.gov) with e-value set as 1e-2 [10]. In parallel, Hidden Markov Model searches were carried out through HMMER v3.1 by matching AAK domain, whose accession number is PF00696.23 in PfamA database [11-13]. The two search results were combined, and screened further with pfam_scan.pl (ftp://sanger.ac.uk/pub/databases/ Pfam) (e_seq: 1e-3, e_dom: 1e-6) [13]. Duplicated and putatively alternative splicing sequences were deleted, which generated a dataset of plant AAK family genes.
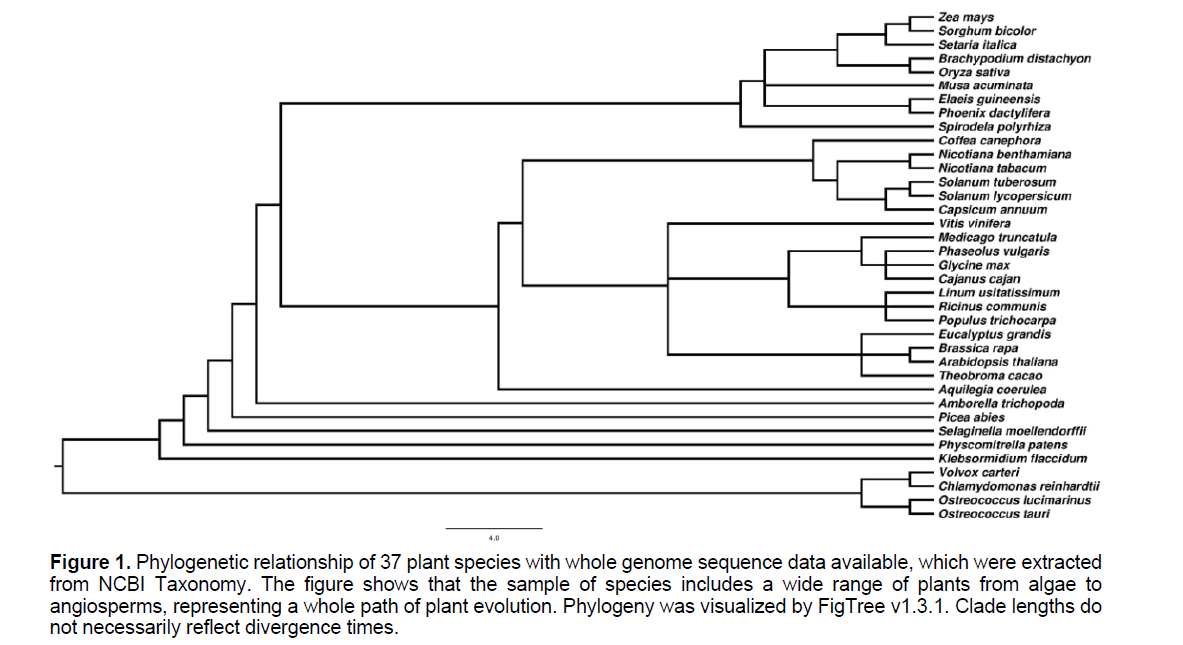
Figure 1:Phylogenetic relationship of 37 plant species with whole genome sequence data available, which were extracted from NCBI Taxonomy. The figure shows that the sample of species includes a wide range of plants from algae to angiosperms, representing a whole path of plant evolution. Phylogeny was visualized by FigTree v1.3.1. Clade lengths do not necessarily reflect divergence times.
2.2 Sequence grouping, alignment and phylogenetic analysis
Dataset of AAK family genes was grouped through OrthoMCL v2.0 using default settings except –I set as 1.5 when running MCL [14]. Alignments were performed using Probcons 1.12 [15]. Phylogenetic analysis was carried out by using PhyML v3.0 under the WAG model [16]. Tree topology was reconstructed using the best of nearest neighbour interchange (NNI) and subtree pruning and regraphing(SPR) methods. Phylogenetic trees were visualized with FIGTREE v.1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
3. Results and Discussion
3.1 Analysis of 64 IPK sequences previously identified
We analyzed all the 64 IPK sequences identified by Dellas et al. as the first step [10]. By using Pfam Scan, which facilitates search of a FASTA file against a library of Pfam HMMs, we identified domains contained in the sequences [8]. As expected, all plant IPK peptide sequences shared an AAK (Amino Acid Kinase) domain (HMM accession number: PF00696.23), whose length was 241 amino acid residues in average [10]. The sequence with the shortest AAK domain was from Montastraeafaveolata, whose AAK domain length was 163 aa residues. All sequences shared a conserved site of histidine residue which was showed to be catalytically essential active site [17]. Although IPK genes are divergent in sequences among the tree of life, it was shown that the 64 plant IPK sequences identified by Dellas et al. [10] formed a monophyletic clade in the phylogenetic tree, which indicated that plant IPKs share a single ancestral IPK sequence.
3.2 Identification of IPK genes from sequenced plant genomes
We performed identification of plant IPK sequences through two steps. Firstly we identified AAK family members comprehensively in sequenced plant genomes, and then extracted IPK genes through further identification and grouping of homologs. To fulfill the first step, we made a comprehensive identification of plant sequences through a combination of approaches of BLAST and HMMER (Materials and Methods). In total, 493 AAK family members were identified from 35 plants with genome sequence data readily available, plus 4 from S. cerevisiae. In the second step, we used OrthoMCL to further identify and group orthologs among AAK family members. In Group 6, 37 sequences were contained, in which Arabidopsis IPK gene (locus ID: AT1G26640.1) was included (Table 1). We back checked these 37 sequences, and were assured that all sequences really had AAK domains.
Table 1. List of IPK genes from sequenced plant genomes
IPK peptide sequence contains a key signature of Histidine residue in IPK peptide sequences (Figure 2) [4,18]. We aligned the 37 peptide sequences, together with two known IPK sequences, from Roseiflexuscastenholzii and Chloroflexusaggregans, respectively, and results showed all 37 peptide sequences have this key His residue (Figure 2). Topology of phylogenetic inference for this group largely conformed to phylogenetic relationship of species where sequences were discovered (Figure 2).
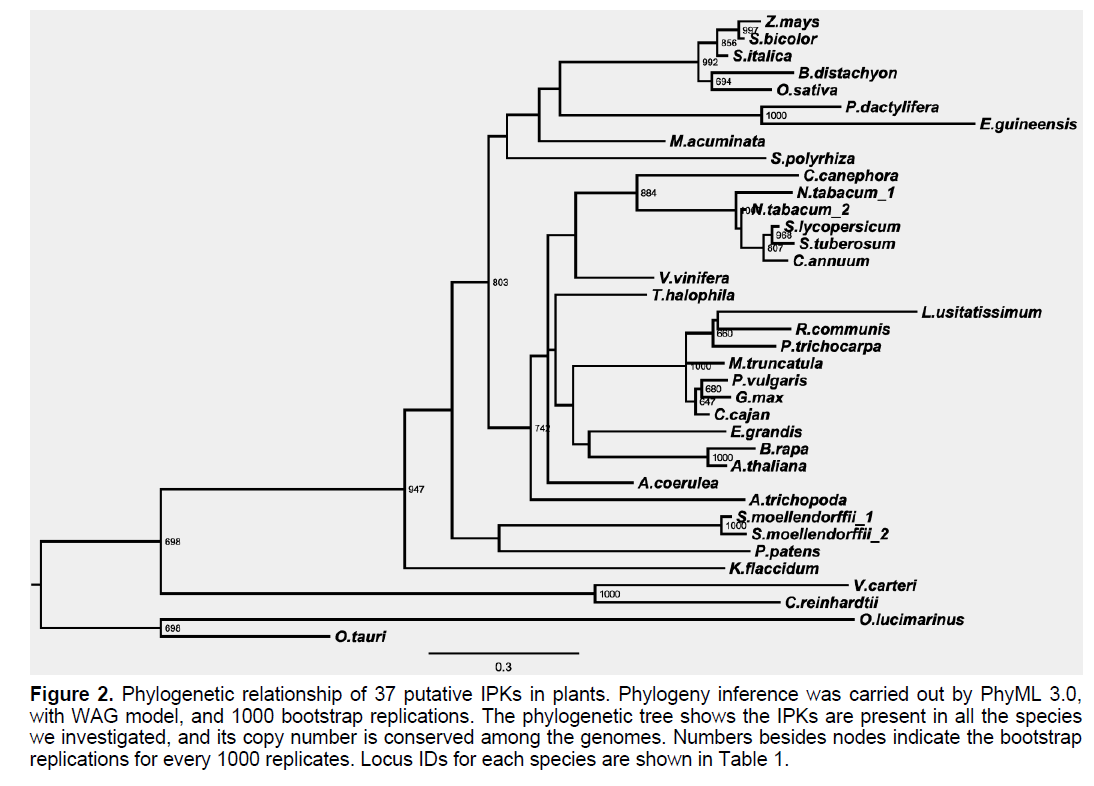
Figure 2:Phylogenetic relationship of 37 putative IPKs in plants. Phylogeny inference was carried out by PhyML 3.0, with WAG model, and 1000 bootstrap replications. The phylogenetic tree shows the IPKs are present in all the species we investigated, and its copy number is conserved among the genomes. Numbers besides nodes indicate the bootstrap replications for every 1000 replicates. Locus IDs for each species are shown in Table 1.
Interestingly, of the 35 plant genomes we checked, every genome has one copy of the putative IPK genes, except for S. moellendorffii and N. tabacum, which contains two, respectively. This suggests important role of IPK gene throughout plant tree of life.
3.3 Phylogeny of AAK family members
In Arabidopsis, 13 AAK family members were identified, among which several were functionally characterized. AT5G13280, AT3G02020 and AT5G14060 were identified to encode aspartate kinases [19-21]. AT1G31230 and AT4G19710 were characterized to have dual activities of aspartate kinase and homoserine dehydrogenase [22]. AT2G39800 and AT3G55610 had delta 1-pyrroline- 5-carboxylate synthase 2 activities [23,24]. AT3G57560 encoded N-acetyl-l-glutamate kinase [25]. 4 AAK members were present in S. cerevisiae, among which 3 were functionally and/or structurally identified. YER052C encoded aspartate kinase, YDR300C encoded enzyme with gamma-glutamyl kinase activity, while YER069W were identified to have acetylglutamate kinase and N-acetyl-gammaglutamyl- phosphate reductase activities [26,27].
We performed phylogenetic inference for these sequences, together with the two IPK sequences from Roseiflexuscastenholzii and Chloroflexusaggregans. AT1G26640 formed a distinctive clade together with the two known IPK genes (Figure 3). No S. cereviaise gene fell into IPK clade, which indicated that no IPK gene was present in S. cereviaise. Functionally identical or similar genes were grouped together, such as YER052C, AT1G31230 and AT4G19710 which encode aspartate kinases.
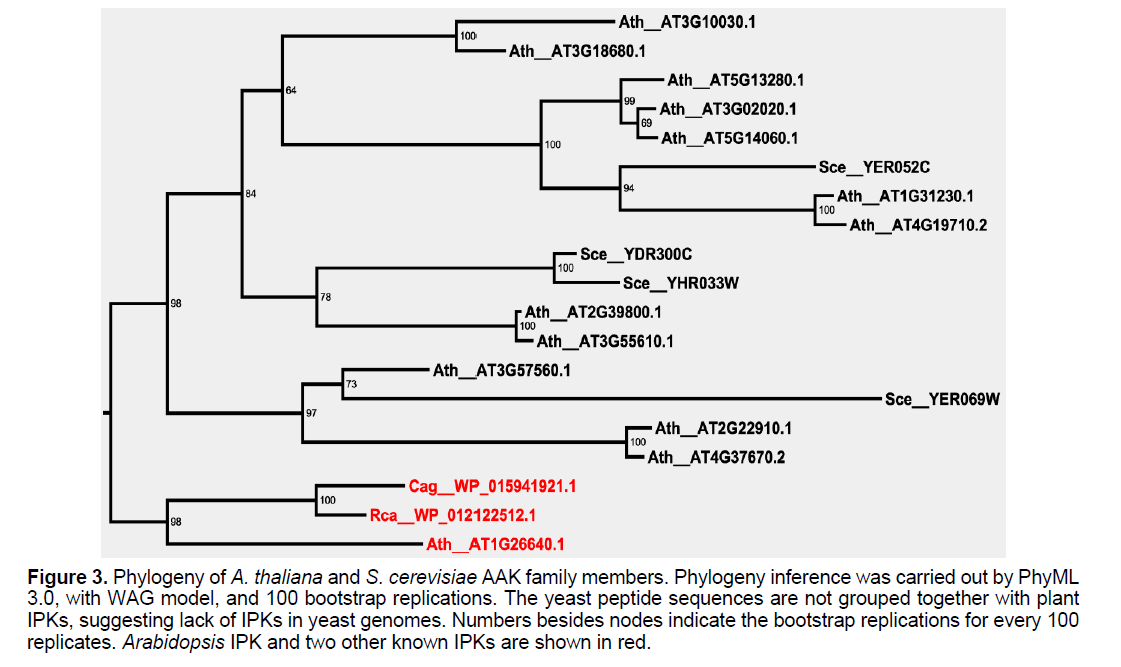
Figure 3:Phylogeny of A. thaliana and S. cerevisiae AAK family members. Phylogeny inference was carried out by PhyML 3.0, with WAG model, and 100 bootstrap replications. The yeast peptide sequences are not grouped together with plant IPKs, suggesting lack of IPKs in yeast genomes. Numbers besides nodes indicate the bootstrap replications for every 100 replicates. Arabidopsis IPK and two other known IPKs are shown in red.
Based on the sequence properties and topology of phylogeny, we concluded that the 37 genes from 35 plant genomes were putative IPK genes. In summary, the genes had His residue signature, and AAK domain; phylogenetic inference showed that Arabidopsis gene AT1G26640 was grouped together with known IPK genes. In evolutionary perspective, plant IPKs had distinctive characteristics. Not like other domains of life, plant IPKs are ubiquitously present in plants, from green algae to higher plants. Copy numbers of IPKs were kept not higher than 2, at least in the 35 plant genomes we checked. Phylogenetic topology of IPK genes and AAK family members indicated that plant IPKs had the same origin which could date back to the emergence of green algae on earth. The results in our report indicated that plant IPKs may have important roles in plants. Further physiological as wells as enzymatic characterization should provide more insights into the role of plant IPKs.
Previous work discovered IPK formed a key point of an alternative route of MVA pathway in different domains of life such as archaea [4,5,7,10]. Here we showed that IPK homologs were present in virtually all plant genomes. Considering that isoprenoids play important roles in plant development and physiology, the IPKs we identified here would shed light on mevalonate pathway of plant isoprenoids biosynthesis. The work we demonstrate here indicates missing points should be filled in characterization of plant isoprenoids biosynthesis network.
Conclusion
In total, 493 AAK family members were identified from 35 plants by combination of BLAST and HMMER. Based on that, 37 putative isopentenyl phosphate kinase (IPK) sequences were identified by OrthoMCL. Analyses including AAK domain analysis and conserved residue identification further confirmed the sequence features of IPK genes. All plant IPK peptide sequences shared an AAK domain (HMM accession number: PF00696.23), and harbored a His residue which is a signature of IPK enzyme. Though IPKs have spotty distribution over the tree of life in other domains of life including animals, fungi, eubacteria and archaea, IPK genes were identified in virtually all plant genomes, and copy numbers of IPKs were very conserved in that no higher than 2 copies remained in all plant genomes. IPKs formed a distinctive clade in phylogenetic tree of AAK gene family. In plant AAK family phylogeny, IPKs formed a distinctive clade, which had a phylogenetic topology conformed to that of plant species.
Acknowledgment
This work was funded by Science and Technology project of Guizhou Tobacco Corporation (Nub. 201704) and the Project of the Tobacco Genomic Program 110201601031 (JY-05).
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Goldstein JL, Brown MS. (1990). Regulation of the mevalonate pathway. Nature. 343: 425-430.
- Rohmer M. (1999). The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat Prod Rep. 16: 565-574.
- Vranova E, Coman D, Gruissem W. (2013). Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Ann Rev Plant Biol. 64: 665-700.
- Lombard J, Moreira D. (2011). Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Mol Biol Evol. 28: 87-99.
- Miziorko HM. (2011). Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch Biochem Biophys. 505: 131-143.
- Boucher Y, Kamekura M, Doolittle WF. (2004). Origins and evolution of isoprenoid lipid biosynthesis in archaea. Mol Microbiol. 52: 515-527.
- Grochowski LL, Xu H, White RH. (2006). Methanocaldococcus jannaschii uses a modified mevalonate pathway for biosynthesis of isopentenyl diphosphate. J Bacteriol.188:3192-3198.
- Mistry J, Bateman A, Finn RD. (2007). Predicting active site residue annotations in the Pfam database. BMC Bioinform. 8: 298.
- Chang WC, Song H, Liu HW, et al. (2013). Current development in isoprenoid precursor biosynthesis and regulation. Curr Opin Chem Biol. 17: 571-579.
- Dellas N, Thomas ST, Manning G, et al. (2013). Discovery of a metabolic alternative to the classical mevalonate pathway. eLife. 2: e00672.
- Camacho C, Coulouris G, Avagyan V, et al. (2009). BLAST+ architecture and applications. BMC Bioinform. 10: 421.
- Eddy SR. (2011). Accelerated Pro file HMM Searches. PLoS Comput Biol. 7: 1-14.
- Finn RD, Bateman A, Clements J, et al. (2014). Pfam the protein families database. Nucl Acids Res. 42: 222-230.
- Li L, Sto eckert CJ, Jr and Roos DS. (2003). OrthoMCL identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178-2189.
- Do CB, Mahabhashyam MS, Brudno M, et al. (2005). ProbCons Probabilistic consistency-based multiple sequence alignment. GenomeRes. 15: 330-340.
- Guindon S, Delsuc F, Dufayard JF, et al. (2009). Estimating maximum likelyhood phylogenies with PhyML. Methods Mol. Biol. 537:113-137.
- Dellas N, Noel JP. (2010). Mutation of archaeal isopentenyl phosphate kinase highlights mechanism and guides phosphorylation of additional isoprenoid monophosphates. ACS Chem Biol. 5: 589-601.
- Matsumi R, Atomi H, Driessen AJM, et al. (2011). Isoprenoid biosynthesis in Archaea - Biochemical and evolutionary implications. Res Microbiol. 162: 39-52.
- Clark TJ, Lu Y. (2015). Analysis of loss-of-function mutants in aspartate kinase and homoserine dehydrogenase genes points to complexity in the regulation of aspartate-derived aminoacid contents. Plant Physiol. 168: 1512-1526.
- Frankard V, Vauterin M, Jacobs M. (1997). Molecular characterization of an Arabidopsis thaliana cDNA coding for a monofunctional aspartate kinase. Plant Mol Biol. 34: 233-242.
- Yoshioka Y, Kurei S, Machida Y. (2001). Identification of a monofunctional aspartate kinase geneof Arabidopsis thaliana with spatially and temporally regulated expression. Genes Gen Syst. 76: 189-198.
- Curien G, Ravanel S, Robert M, et al. (2005). Identification of six novel allosteric effecttors of Arabidopsis thaliana aspartate kinase-homoserine dehydrogenase isoforms. Physiological context sets the specificity. J Biol Chem. 280: 41178-41183.
- Leprince AS, Magalhaes N, de Vos D, et al. (2014). Involvement of Phosphatidylinositol 3- kinase in the regulation of proline catabolism in Arabidopsis thaliana. Front Plant Sci. 5: 772.
- Szekely G, Abraham E, Cseplo A, et al. (2008). Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 53: 11-28.
- Chen YM, Ferrar TS, Lohmeier-Vogel EM, et al. (2006). The PII signal transduction protein of Arabidopsis thaliana forms an arginine-regulated complex with plastid N-acetyl glutamate kinase. J Biol Chem. 281: 5726-5733.
- Rafalski JA, Falco SC. (1990). Structure of the yeast HOM3 gene which encodes aspartokinase J Biol Chem. 265: 15346.
- Li W, Brandriss MC. (1992). Proline biosynthesis in Saccharomyces cerevisiae molecular analysis of the PRO1 gene which encodes gamma-glutamyl kinase. J Bacteriol. 174: 4148-4156.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences
