Genomic Analysis of fgf in zebra Fish (Danio Rerio)
Ayesha Tahir*
Ayesha Tahir*
Department of Bio Science, comsats University, Islamabad Campus, Pakistan
- Corresponding Author:
- Ayesha Tahir
Department of Bio Science, comsats University
Islamabad Campus, Pakistan
Tel: 03447726566
E-mail: ayeshatahir193@gmail.com
Received date: April 19, 2021; Accepted date: May 04, 2021; Published date: May 11, 2021
Citation: STahir A (2021). Genomic Analysis of fgf in zebra Fish (Danio Rerio). Electronic J Biol 17(S3): 213 -217
Abstract
FGFs are abbreviated as “Fibroblast growth factors”. These "FGFs" are the "secreted “development factors of polypeptide. The "signaling" scheme of FGFs is actually the vital part which "plays" a significant role in many processes of development in vertebrates. The humans also have FGFs family. This family comprises of 22 members. On the other hand, zebra fish family has 16 members while eleven more zebra fish fgf were identified by homo-logy based search. This search was conducted in zebra fish genome and cDNA databases. By calculating additional member’s fgf family of zebra fish now consist of 27 members at least. By the conduction of polygenetic processes and gene location analyses, we identified relationships between human FGFs genes and zebra fish Fgf genes. This paper also evaluates and represents the phylogenetic information of fgf zebra fish family.
Keywords
Fgf, zebra fish, Phylogenetic analysis, Human fgf, Gene location, Fibroblast
Introduction
(FGFs) are actually the development factors named as "Fibroblast development factors”. They structure a group of commonly "extracellular" flagging "peptides", which are the controllers of various "organic" cycles going from "cell-multiplication" to the command of undeveloped advancement in "metazoans" [1]. Most FGF qualities are discovered dissipated all through the "genome". In human, "22 FGF” qualities have been recognized and the chromosomal areas of all with the exception of FGF16 are known. A few human FGF qualities are bunched inside the "genome" [2]. “FGF3, FGF4 and FGF19” are situated on “chromosome 11q13” and are isolated by just forty and ten kb, separately; “FGF6" and " FGF23” are situated inside fifty kb on" chromosome" [3] “12p13; and FGF17 and FGF20” guide to “chromosome 8p21-p22”. These quality areas demonstrate that the "FGF-qualityfamily" was produced by "quality" and "chromosomal" duplication and movement throughout advancement. [4] Curiously, a transcriptionally dynamic segment of human FGF7, situated on chromosome “15q13-q22”, has been intensified to around 16 duplicates, which are scattered all through the human genome [4]. "FGFs" are indicated in the two "vertebrates" and "spineless" creatures [5]. Various "FGF" qualities are distinguished in "vertebrates", incorporating 10 "FGFs" in zebra fish “FGF2–4, 6, 8, 10, 17a, 17b, 18, 24”, 6 in "Xenopus"“FGF2–4, 8–16", thirteen in "chickens" “FGF1–4, 8–10, 12, 13, 16, 18–22", 22 in "mice" "FGF1–18, 20–23" and people "FGF1–14, 16– 23”, though just three "Drosophila" "FGF" qualities [6] and two "Caenorhabditis" elegans "FGF “qualities have been seen in "spineless" creatures [7]. Human "FGFs" contain twenty-two individuals: “FGF1, FGF2, FGF3 (INT2), FGF4, FGF5, FGF6, FGF7 (KGF), FGF8 (AIGF), FGF9, FGF10, FGF11, FGF12, FGF13, FGF14, FGF16, FGF17, FGF18, FGF19, FGF20, FGF21, FGF22, and FGF23” [8]. The FGF family contains twenty-three individuals, despite the fact that there is just eighteen “FGFR” ligand. Four relatives don't tie with "FGFR" as "FGF" "homologous “elements “(FGF11, FGF12, FGF13, and FGF14)” [9] and are all the more accurately alluded to as "FGFhomologous- variables" [10]. Furthermore, there is no human "FGF15" quality; the quality "orthologous"to "mouse" "FGF15" is "FGF19” [11].By "phylogenetic” examination, the "human FGF" quality family can be separated in to 7 sub families: “FGF1, FGF4, FGF7, FGF8, FGF9, FGF11, and FGF19. The FGF1, FGF4, FGF7, FGF8, FGF9, FGF11, and FGF19 sub families include FGF1 and 2, FGF4, 5, and 6, FGF3, 7, 10, and 22, FGF8, 17, and 18, FGF9, 16, and 20, FGF11, 12, 13, and 14, and FGF19, 21, and 23”, separately [12]. As opposed to "phylogenetic" investigation, quality area examination shows that the human FGF quality family can be isolated in to 6 sub families: “FGF1/2/5, FGF3/4/6/19/21/23, FGF7/10/22, FGF8/17/18, FGF9/16/20, and FGF11/12/13/14” [13]. Individuals from the “FGF8, FGF9, and FGF11” sub-families are familiar to those of the “FGF7/10/22, FGF8/17/18, FGF9/16/20, and FGF11/12/13/14” sub families in the quality area investigation [14]. During undeveloped turn of events, FGFs have assorted jobs in directing cell expansion, relocation and separation [15]. In the grown-up life form, FGFs are homeostatic factors and capacity in tissue fix and reaction to injury. At the point when improperly communicated, some FGFs can add to the pathogenesis of malignant growth [16]. A subset of the FGF family, communicated in grown-up tissue, is significant for neuronal sign transduction in the focal and fringe sensory systems. FGFs are little proteins (somewhere in the range of 17 and 34 kDa) described by a generally very much preserved focal space of “120 to 130 amino acids" [17]. This area is coordinated into twelve “antiparallel" "β" sheets shaping a three-sided substance called "beta-trefoil" [18] When all is said in done, FGFs work through restricting to a "tyrosine-kinase-receptor" "FGFR" on the outside of the cell film. Two "FGF-ligands" tie a "dimeric-receptor" within the sight of "heparin-sulfateproteoglycan” permitting the "transphosphorylation" and attachment of the "intracellular-tyrosine-kinase “space of the receptor [2]. Through the initiation of the "cytoplasm" pathways, the “FGF-signal-controls" a few significant cell capacities like cell expansion, relocation, separation, or endurance [9].
Material and Method
Identification of fgf genes in zebra fish
PSI Blast, protein blast [BLAST P] and Delta Blast were performed in zebra fish to regain the protein and cDNA sequence of fgf genes. After it, by using the different databases, the target sequences were processed. For example: Pfam databases, smart databases, and NCBI [conserved] data bases. cDNA sequences, chromosomal location, length of protein and their genomic information [genomic information related to the fgf genes] were taken from NCBI.
Gene structure and prediction:
Gene structure display server is a freely online available Server. Schematic representation and gene structure analysis is done by this server.
Gene location analysis
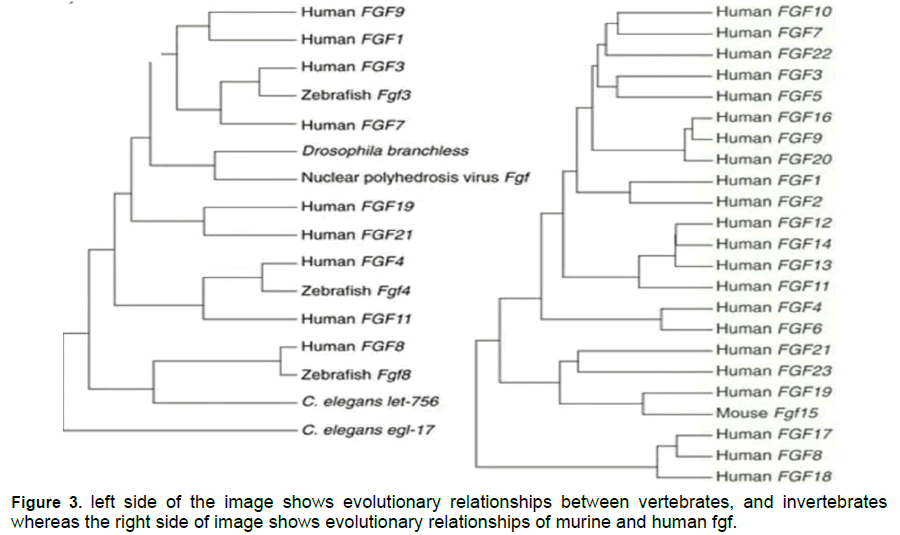
Ensemble genome browser (ensembl.org) is a browser by which the positions of "human" and "zebra fish" "genome" on "chromosomes" are obtained. However, “psychogenetic" analyses and "gene located" analyses pointed the potential evolutionary relationship. However, gene evolutionary relationships can never be determined only by it. However, the "psychogenetic" analyses and gene located analyses also indicate potential evolutionary relationship in gene formation [19]. At human "fgf7"and "fgf 22" loci and zebra fish "fgf7"and "fgf22" loci, conserved gene orders were examined. They indicate that zebra fish "fgf7" and "fgf22" are orthologues of human "fgf7" and "fgf22”. At human "fgf10" and "fgf10a" loci a "conserved" gene- order was obtained. It points that "fgf10a" is a "zebra fish" of "human-fgf10"
Phylogenetic analysis
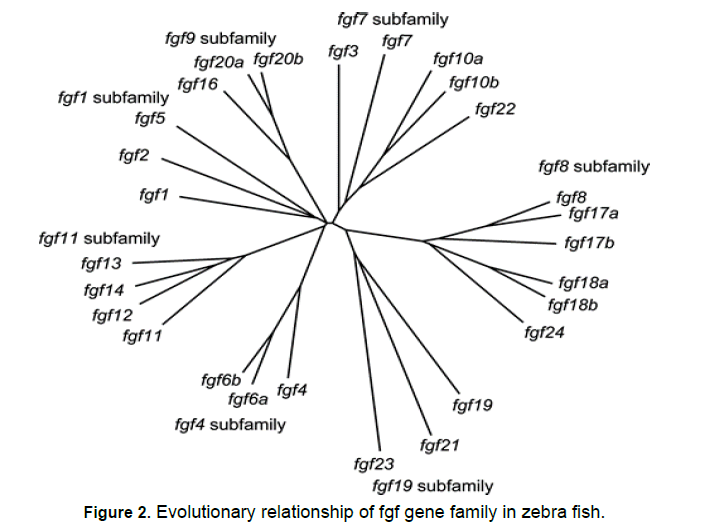
An online software tool named as mega 7 was used to make Fgf genes in zebra fish of polygenetic analysis. it is accessed from(https://mega.software.informer. com/7.2/) However "polygenetic analysis" points that human “Fgf gene family” are further termed in to sub families. Such as “FGF1, FGF4, FGF7, Fgf8,Fgf9 and Fgf11”. These subfamilies consist of “Fgf1=2, fgf4=5, fgf3=7, fgf10=22, fgf8=17=18, fgf9=16=20, fgf11=12=13=14 and fgf19=21=23” by order. On the contrary, "zebra fish" "fgf-gene-family" are also further termed in to sub families. Zebra fish "fgf" sub families are potentially coherent with the human fgf families. Human "FGF5" is a fellow member of the “FGF4” sub family, and “zebra fish fgf5” is a member of the fgf1 subfamily.
Results
Phylogenetic analysis
Figure 1 to 3
Discussion
FGFs abbreviated as fibroblast growth factors. The humans also have FGFs family. This family comprises of 22 members [20]. On the other hand, zebra fish family has 16 members while eleven more zebra fish fgfs were identified by homo-logy based search. This search was conducted in zebra fish genome and cDNA databases. By calculating additional members FGF family of zebra fish now consist of 27 members at least [21]. By the conduction of "polygenetic “processes and "gene location analyses “we pointed relationships between human "fgf-genes" and zebra fish "Fgf-genes" [22]. After the research, we found that there were no extra undefined fgf. During the evolution, it might be possible that fgf9 has been lost. In zebra fish fgf family, there are six paralog [23]. If there is a comparison of mammalian gene with Teleost fish, teleost was shown in it. Including zebra fish there are more often to homo- logos of mammalian equivalent. It proposed that that shortly after the teleost radiation, there is additional genome duplication [24]. This may be partially or wholly genome duplication. It is preceded by rapidgene- loss. It is because gene "duplication" works only for the 20% of zebra fish genes identity. By the process of gene duplications, zebra fish fgf paralog were generated. This whole process is done by the location analyses of zebra fish fgf genes [25]. Fibroblast development factors (FGFs) are a group of fundamental factors that control creature advancement and physiology; in any case, their job in cetacean advancement isn't obviously perceived. Here, we sequenced the blade whale genome and dissected FGFs from 8 cetaceans. FGF22, a hair follicle-advanced quality, displayed pseudogenization, demonstrating that the capacity of this quality is not, at this point fundamental in cetaceans that have lost the majority of their body hair. A transformative investigation uncovered marks of positive determination for FGF3 and FGF11, qualities identified with ear and tooth advancement and hypoxia, separately. We discovered a D203G replacement in cetacean FGF9, which was anticipated to influence FGF9 homodimerization, recommending that this quality assumes a part in the procurement of inflexible flippers for productive moving. Cetaceans use low bone thickness as a lightness control system, yet the fundamental qualities are not known. We tracked down that the declaration of FGF23, a quality related with diminished bone thickness, is extraordinarily expanded in the cetacean liver under hypoxic conditions, subsequently embroiling FGF23 in low bone thickness in cetaceans. Out and out, our outcomes give novel bits of knowledge into the parts of FGFs in cetacean variation to the oceanic climate.
Furthermore, the "phylogenetic analyses “and "gene located analyses" pointed that orthologues of "fgf6b"and "fgf6a"are the "paralog" of "fgf6b". "fdf10a"was basically examined as "orthologues" of human "fgf10". "Fgf10b" is a "paralog “of ‘’fgf10’’ [26]. Zebra fish "fgf17a"was examined as "orthologues" of "fgf17a". "Fgf17a" is actually the Para log of "fgf8". "Fgf24"was actually affiliated to "fgf8" as "paralog" of zebra fish. [27]. The "phylogenetic Analyses “and "gene located analyses" pointed that auto log of "fgf24"is a Para log of "fgf18a" but not a Para log of "FGF8"it means that "fgf18a"has two paralog which are fgf18b and fgf24 [28] Fgf20a was examined as zebra fish orthologues of" fgf20" However, further studied shows that orthologues of “fgf20b and fgf20a’’ are paralog of "fgf20b" [29]. At human Fgf 18 locus a gene order was ascertained. Their gene order was actually conserved. While the same "conserved "gene order was also observed at zebra fish fgf18b [30]. However, the same gene order was also studied at fgf 24 loci. They all were latently yielded from same ancestor gene [31].They were yielded by the duplication of genes. "FGF18" and "FGF-24" were nearly placed to each other on “chromosome 14”. According to "phylogenetic" analysis, human "fgf 18" is more nearly affiliated to zebra fish "fgf 24". From the above analyses shows that fgf 24 is paralog of "fgf- 18a" [32]. It is also discussed above that they are latently yielded by the process of local gene duplication [33]. While On the contrary, it is ambiguous from the gene location analyses that which of them “fgf18a or Fgf18b” is a zebra-fish "orthologues" of the “human-fgf18" [34- 36] fgf10a, fgf16, and fgf24 are needed for the ï¬ÃÂn bud to shape. Fgf19 is needed for forebrain development. Fgf20a is needed for starting ï¬ÃÂn regeneration. Fgf21 is needed for hematopoietic .As zebra ï¬ÃÂsh undeveloped organisms are little, the preparation and resulting early stage advancement happen remotely, and the undeveloped improvement is fast.
Conclusion
The current study indicates that Human "fgf 18" is more closely related to zebra fish "fgf 24" according to "phylogenetic" study. According to the results of the above studies, fgf 24 is a paralog of "fgf- 18a" They are also latently generated by the method of local gene duplication. it is unclear whether “fgf18a or Fgf18b” is a zebra-fish "orthologue" of the "human-fgf18"fgf10a, fgf16, and fgf24 are needed for the fin bud to form based on gene location analyses. Fgf19 is required for the development of the forebrain. Since the undeveloped species of zebra fish are small, the planning and subsequent early stage development take place remotely, and the undeveloped development is rapid.
References
- Akter T (2009). Climate change and flow of environmental displacement in Bangladesh. Centre for Research and Action on Development, Dhaka.
- Ahmad M (2005). Living in the coast: urbanization. Program development office for integrated coastal zone management plan project, water resource planning organization.
- Buvinic M, Gabriela V, Mauricio B (1999). Hurricane Mitch: Women's Needs and Contributions Inter-American Bank 4:363.
- Byravan S, Rajan SC (2009).The social impacts of climate change in South Asia. J Refug Stud 5:34-147.
- www.weadapt.org/knowledge-base/wikiadapt/bangladesh-ncap-proj
- Berner J, Furgal C (2004). Human health. Climate Change: Impacts, Adaptation and Vulnerability Sci Rep 14:391-431.
- Banu D. (2002). BRAC-char development and settlement project 2000 - 2004. Research Reports 30:476–512.
- Hossain MA, Reza MI, Rahman S, et al. (2012).Climate change and its impacts on the livelihoods of the vulnerable people in the southwestern coastal zone in Bangladesh. In Climate change and the sustainable use of water resources 237-259.
- Boano C, Zetter R, Morris T (2012). Environmentally displaced people: Understanding the Linkages between Environmental Change, Livelihoods and Forced Migration.
- Douglas W, Pherson M (2008). Global Climate Change and Extreme Weather Events: Understanding the Contributions to Infectious Disease Emergence: Workshop Summary. Proc Natl Acad Sci USA.
- Faruque A (2007). Resettlement policy resettlement policy development: The case of development: The case of Bangladesh. Asian Development Bank Bangladesh, Resident Mission, Power Point Presentation.
- Ginexi EM, Weihs K, simmens SJ, et al. (2008). Natural disaster and depression: a prospective investigation of reactions to the 1993 mid west floods. J Community Psychol 28: 495-518.
- Hartmann B, Barajas-Roman E (2009). Reproductive Justice, Not Population Control: Breaking the Wrong Links and Making the Right Ones in the Movement for Climate Justice. In WE ACT for Environmental Justice conference on Advancing Climate Justice: Transforming the Economy, Public Health and Our Environment 29-30.
- Hovey JD, Magana C (2000). Acculturative stress, anxiety, and depression among Mexican immigrant farm workers in the Midwest United States. J of immi heal 2:119-31.
- Paradies Y (2006). A review of psychosocial stress and chronic disease for 4th world indigenous peoples and African Americans. Ethn Dis 16: 295.
- Byravan S, Rajan SC (2009). The social impacts of climate change in South Asia. J of Migr and Refu Issues 5:134-47.
- Nishat A, Hussein SG, Matin MA et al. (2009) Adapting to Climate Variability and Change in Bangladesh.
- Cheatham AR, Paredes DG, Griffin S et al. (2009). Looking both ways: women’s lives at the crossroads of reproductive justice and climate justice. Asian Communities for Reproductive Justice 5: 1-30.
- Rehner TA, Kolbo JR, Trump R, Smith C et al. (2000). Depression among victims of south Mississippi's methyl parathion disaster. Health & social work 25:33-40.
- Satterthwaite D (2009).The implications of population growth and urbanization for climate change. Environment and Urbanization. 21: 545-567.
- http://ptsd.about.com/od/causesanddevelopment/a/EffectofKatrina.html)
- Viswanathan S, Srivastava R (2007). Learning from the Poor: Findings from Participatory Poverty Assessments in India. Asian Development Bank.
- Choudhury WA, Quraishi FA, Haque Z (2006). Mental health and psychosocial aspects of disaster preparedness in Bangladesh. International review of psychiatry 18:529-35.
- http://ptsd.about.com/od/causesanddevelopment/a/EffectofKatrina.html)
- Cameron L, Shah M (2015. Risk-taking behavior in the wake of natural disasters. J Hum Resour 50:484-515.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences