Genetic Variation and Phylogenetic Relationships of Safflower using Morpho-Phenological Markers
Moslem Bahmankar, Daryoosh Ahmadi Nabati, Masood Dehdari, Seyyed Hamid Reza Ramazani
Moslem Bahmankar1*, Daryoosh Ahmadi Nabati2, Masood Dehdaris3, Seyyed Hamid Reza Ramazani4
1Department of Agronomy and Plant Breeding, College of Aburaihan, University of Tehran, Tehran, Iran;
2Department of Agronomy and Plant Breeding, College of Agriculture, University of Shahid Chamran Ahvaz, Ahvaz, Iran;
3Department of Agronomy and Plant Breeding, College of Agriculture, University of Yasuj, Yasuj, Iran;
4Department of Agronomy and Plant Breeding, College of Agriculture, University of Birjand, Birjand, Iran.
Received Date: August 09, 2016; Accepted Date: September 23, 2016; Published Date: September 30, 2016
Citation: Bahmankar M, Nabati DA, Dehdari M, et al. Genetic Variation and Phylogenetic Relationships of Safflower using Morpho-Phenological Markers. Electronic J Biol, S:1
Abstract
Safflower (Carthamus tinctorius L.) is an important oilseed crop which is cultivated predominantly in semiarid and temperate regions of the world. In order to assessment of genetic variation, relationships between traits and phylogenetic clustering of genotypes, an investigation was conducted as a randomized complete block design with three replications and 20 safflower genotypes. The results of analysis of variance showed the significant differences among of genotypes for total of the studied traits including days to flowering, days to maturity, plant height, head per plant, seeds per head, main head diameter, 1000 seeds weight and seed yield per plant. Soviet union-1 genotype possessed the greatest values of total traits especially seed yield per plant that ranked this genotype as superior genotype. A high correlation coefficient (r=0.80*) was obtained between 1000 seeds weight and seed yield per plant. Cluster analysis classified genotypes into three distinct groups. Results of F Beele's type statistic showed that there was a significant difference between groups for all the studied traits except days to maturity, plant height and heads per plant. Generally, results indicated that clustering some foreign genotypes into Iranian group was due to their similar genetic basis and as well as it was suggested that Iran may be the origin of them.
https://betsatgirisi.com https://bettilte.com https://vegabete.com https://kanyongiris.com https://matgiris.com https://celtabetegiris.com https://hilbetegiris.com https://melbete.com https://kinbettinge.com https://wipbett.com https://pusulabetegiris.com https://superbahiss.com https://lidyagiris.com https://holiganbete.com https://1xbetgiriss.com https://asyabahise.com https://jetbahise.com https://betdoksana.com https://betebetle.com https://betgramagiris.com
Keywords
Safflower; Cluster analysis; F Beele's type; Correlations coefficients.
Introduction
Safflower (Carthamus tinctorious L.) is an annual and diploid2 )n=2x=24( plant belong to composite family [1,2]. Middle East and Iran are origins of safflower [1,3]. It is used mainly in production of cooking oil with unsaturated fatty acids [2,4]. Genetic variation has an important role in plant breeding because of the hybrids obtained from lines with more genetic variation showing more heterosis than the convergent races [5]. In plant species, genetic variation occurs in many ways such as mutation, generic recombination, migration and gene flow, genetic derivation, interspecies hybrid, polyploidy and selection [6]. Iran is one of the main regions of safflower genetic resources and the cultivation of this plant in Iran is faced with some limitations because of lack of environmental stressresistant and improved varieties [7]. As regards to the lack of sufficient breeding programs for this plant, applying researches on breeding and production of modified genotype to the region is very important [8]. In a research, 66 lines of safflower were studied for genetic variation of different agronomical traits such as the number of days to germination, the number of days to flowering, 50 percent of flowering and maturity, plant height, seed yield and components of seed yield. The results show that there was a significant difference between investigated lines for all traits and genotypes are located in three groups which have significant differences in view of all the traits except in the number of days to germination [8]. The results of other studies show that there were significant differences between genotypes for agronomical traits [6,9]. Correlation among traits is important to execute the exact breeding programs [10]. Correlation coefficients were used to determine the relationship between yield and its components and also the relationships between yield components in safflower [10-12]. It is reported that there was a positive and significant phenotypic and genotypic correlation between seed yield and other traits including number of heads per plant, head weight, number of seeds per head and 100 seeds. Also the results show that the maximum phenotypic and genotypic correlation was observed between the two traits of number of head per plant and seed yield [13]. In another study on different traits of safflower, it is mentioned that 1000 seeds weight had the maximum correlation with seed yield [11,12]. According to the fact that Safflower is a native oil seed plant of Iran and there was no study on the genetic variation of safflower in Yasooj condition, so, we conduct this research to recognize the relationships between different traits and as well as select the superior genotypes in order to use them in safflower breeding programs.
Materials and Methods
Plant materials and experimental design
Twenty safflower genotypes (Table 1) including; 6 Iranian genotypes and 14 exotic genotypes (provided from Institute of Plant Genetics and Crop Plant Research (IPK) in Gatersleben of the Germany) were cultivated at research farm of the Researches Center for Agriculture and Natural Resources with a loam soil, Yasuj (51º 31´ E and 30º 41´ N, 1734 m asl) in 2013.
| Origin | Genotype | Origin | Genotype | Origin | Genotype |
|---|---|---|---|---|---|
| Polish | C19 | Morocco | C130 | Iran | Khatam Yazd |
| Soviet Union | C160 | Germany | C132 | Iran | Goldasht |
| Republic of Czech | C9 | Soviet Union | C161 | Iran | Sina411 |
| USA | C56 | Morocco | C24 | Iran | Local Isfahan |
| Iran | IL111 | Pakistan | C151 | Iran | Isfahan 14 |
| Polish | C55 | Tajikistan | C83 | Indian | C173 |
| Germany | C159 | Pakistan | C124 |
Table 1: Safflower genotypes collected from different location.
A randomized complete block design with three replications was used. Each plot consisted of four rows with 3 m long and 50 cm apart spaced. Seeds were sown by hand with 5 cm distance in rows. Recommended crop management practices were implemented to raise the crops to desirable growth stages. Plots were kept weed, pest and disease free till harvesting time.
Studied traits and their measurement
Regarding the marginal effects 6 to 8 plants were selected randomly and measured all of phenological and morphologic traits include days to initiate flowering (DIF), days to maturity (DM), plant height (PH), head per plant (HP), main head diameter (MHD), seeds per head (SH), 1000 seeds weight (SW) and a seed yield per plant (SYP).
Statistical analysis
Analyses of variances (ANOVA) were carried out using PROC GLM of SAS [4]. Mean comparisons were conducted using Duncan method. Components of variance including genetic variance and environmental variance determined according to mathematical expectation of mean of squares. Then the genetic standard deviation was estimated using square root of genetic variance. Genetic covariance of two traits was estimated using obtained information from Multivariate Analysis of Variance of randomized complete block design and SAS software via the following relationship (1):
Covg = (sscpv / dfv sscpe / dfe) / r − − − − − − − − − − − − −− > (1)
In this relation, sscpv obtained from sum of squares of products variety matrix, sscpe obtained from sum of squares of products error matrix, dfv and dfe denote the freedom degree of varieties and freedom degree of error, respectively. As well as in the relation 1, r denotes the number of replication. Phenotypic and genetic correlation coefficients calculated in order to investigate the phenotypic and genotypic relationships between yield and its components. Phenotypic and genetic correlation coefficients were calculated by the following relationship [14,15]:
rp(x,y) = covp (x,y) / (sx,sy) − − − − − − − − − − − − − − > (2)
rg(x,y) = covg (x,y) / (sx,sy) − − − − − − − − − − − − > (3)
In the above relations (2 and 3), covp and covg denotes the phenotypic and genetic covariance of x and y traits and sx and sy are genetic standard deviations of these two traits, respectively. Clustering of the similar genotypes based on measured traits was done by Ward method cluster analysis according to Euclidean distance square. Logical number of groups obtained from cluster analysis was determined by dendrogram and F Beele's type statistic test [16]. Groups in above mentioned statistic test considered as treatment and individuals inside them implied as replication (completely random unbalanced design). After variance analysis if F is significant for most of measured traits (more than 50%) it is inferred that the number of groups was selected correctly [16].
Results and Discussion
Analysis of variance
Results of ANOVA showed that there is a significant difference between genotypes for all studied traits including days to initiate flowering, days to maturity, plant height, head per plant, main head diameter, seeds per head, 1000-seeds weight and a seed yield per plant (Table 2). These differences represent the existence of variation among genotypes based on studied traits and these differences can be used in breeding programs. In another study on safflower, there was a significant difference between studied genotypes in view of agronomical traits [6]. In this research, maximum amount of phenotypic and genetic variances among evaluated traits observed for the main head diameter (Table 2). Non-significant difference of phenotypic and genetic variances of main head diameter trait indicated that most of the observed variation for this trait caused by genetic factors and therefore selection can be effective for the main head diameter trait (Table 2). Genetic variation among different genotypes of safflower in view of these agronomical traits was reported in other studies [8,9].
| Mean of Square | Source | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| SYP | MHD | SW | SH | HP | PH | DM | DIF | df | |
| 5.87 | 40.8* | 0.44 | 6.85 | 0.74 | 0.78 | 24.4** | 11.3 | 2 | Replication |
| 47.5** | 5650** | 1118.3** | 79.9** | 8.1** | 262.1** | 113.1** | 93.5** | 19 | Genotype |
| 0.2 | 10.7 | 0.2 | 2.2 | 1.1 | 1.4 | 2.6 | 4.7 | 38 | Error |
| 22.2 | 1.4 | 1.3 | 4.3 | 9.4 | 1.7 | 1.4 | 3.5 | CV (%) | |
| 13.34 | 39.36 | 1879.8 | 46.21 | 3.88 | 86.87 | 36.82 | 29.6 | - | Genetic Variance |
| 20.74 | 39.55 | 1891.95 | 46.9 | 6.83 | 88.28 | 40.56 | 34.66 | - | Phenotypic Variance |
DIF: Days to Initiate Flowering; DM: Days to Maturity; PH: Plant Height; MHD: Main Head Diameter; HP: Heads Per Plant; SH: Seeds Per Head; SW: 1000-Seed Weight; SY: Seed Yield Per Plant
Table 2: Results of analysis of variance of the nine measured traitsof 20 safflower genotypes.
Mean comparisons
The results of mean comparisons of the studied genotypes are shown in Table 3. There was variation between genotypes for days to maturity. Isfahan-14 genotype with 134.7 days to maturity and India-2 genotype with 105 days to maturity were serotinous and precocious genotypes, respectively (Table 3). Variation of maturity period of different safflower genotypes was reported in another study and it was concluded that precocious genotypes are very important for the regions with limited growing stresses in end of growing season [6,13]. Minimum and maximum heights were observed in Morocco-2 and Esfahan-14 genotypes, respectively. So, Isfahan-14 genotype was recognized as the highest genotype in this study (Table 3). Considering this fact that strong blow of wind can be a limitative factor of cultivating tall genotype and since these genotypes are not suitable for mechanized harvesting operation, so it should concentrate on genotype with adequate plant height (60–80 cm). Therefore, according to the significant difference between genotypes for plant height, it is possible to select suitable genotypes for mechanized harvest. The weight of 1000seeds ranged from 26/5 g for Pakistan genotype to 46/4 g for Soviet genotype (Table 3). Other studies reported the smaller range of weight of 1000-seeds [12]. In this research maximum and minimum seed yield per plant belong to Soviet-1 genotype with 21/9 g and Poland-2 genotype with 6/9 g, respectively. Also, another study showed significant difference between safflower genotypes for this trait [6]. Soviet union-1 genotype produced higher grain yield and ranked as the superior genotypes.
| Genotypes | DIF | DM | pH(cm) | HP | SH | SW(gr) | MHD(mm) | SYP(gr) |
|---|---|---|---|---|---|---|---|---|
| Yazd | 56.6 gh | 109 i | 61 h | 11bcd | 34.2 def | 40.8 e | 256 cd | 15.2 bcd |
| Goldasht | 58.3 fgh | 120cd | 63 h | 9.5 de | 42.5a | 37.5fg | 297 a | 13.5 c -g |
| Sina411 | 54.7h | 117 d | 72f | 9.4 de | 32 ef | 45.1b | 261c | 14.3 cde |
| Local Isfahan | bc68.3 | 119 cd | 75 e | 13.4 a | 39 b | 30.2 j | 244 fg | 13.4 c -f |
| Isfahan14 | 72 b | 134 a | 96a | 13.6 a | 35 cde | 35.3 h | 251 de | 17.8 abc |
| Soviet Union1 | 66.6c | 112 fgh | 85 b | 14 a | 37 bcd | 46.4 a | 293a | 21.9 a |
| German1 | 65.3 cd | 115ef | 72 f | 9.3de | 34.5 def | 29k | 185 k | 8.6 fgh |
| Soviet Union2 | 60 efg | 118 cd | 82 c | 11 bcd | 38 b | 35.6 h | 278 b | 15 bcd |
| Morocco 1 | 57.3 gh | 110 hi | 77de | 11 b-e | 37.6 bc | 35.5 h | 243 g | 12.6 c -g |
| Morocco 2 | 60 efg | 111ghi | 63h | 10.6 b-e | 37 bcd | 32.3 i | 195 j | 10.5deh |
| Polish1 | 60.6efg | 118 de | 66 g | 11.6 bc | 28g | 32.2i | 186 k | 9.5 e -h |
| IL111 | 62.6de | 121bc | h | 12.4 ab | 35cde | 43.5 c | 283b | 19.5 ab |
| Indian1 | 77 a | 118cd | 81 c | 9 e | 25 h | 28.2 kl | 174 m | 8.5 fgh |
| Pakistan | 59efg | 114f | 75 e | 10.5 b -e | 37 bc | 26.2 n | 205 i | 9.7 e -h |
| Indian2 | 62.6de | 105 j | 78 d | 11 b -e | 32f | 27.3 m | 179l | 8.4 gh |
| Tajikstan | 62 def | 123 b | 65 g | 9 e | 44.5 a | 38.2f | 293a | 12.4 d -g |
| Czech | 67 c | 114 f | 67 g | 10 cde | 27gh | 37.2 g | 184 kl | 11.3 d -h |
| America | 69 bc | 119cd | 62 h | 9.5 de | 28.6g | 42.4 d | 249ef | 12.8 c-g |
| Polish2 | 60 efg | 114fg | 65 g | 10.6 b-e | 39 b | 27.8 lm | 228 h | 6.9 h |
| German2 | 65 cd | 115ef | 72f | 13.5a | 28.6g | 30.1 j | 184kl | 8.5fgh |
DIF: Days to Initiate Flowering; DM: Days to Maturity; PH: Plant Height; MHD: Main Head Diameter; HP: Heads per Plant; SH: Seeds per Head; SW: 1000-Seed Weight; SYP: Seed Yield per Plant
Table 3: Mean Comparisons ofstudied genotypes for measured traits in safflower.
Correlation coefficients analysis
The results obtained from calculating phenotypic and genetic correlation coefficients are shown in Table 4. Results showed a positive and high correlation between traits of days to initiate flowering and days to maturity (Table 4). This relationship shows that genotypes with earlier flowering have a shorter growing period and are more precocious. So it seems that selection for flowering phase is effective for precocity maturity. This high correlation are reported in another study [12]. Correlation between plant height and heads per plant was positive and significant (Table 4). This study revealed that taller genotypes produce more heads, so, it is inferred that potential of producing head in genotypes increased with increase in plant height (Table 4). A positive and significant correlation observed between plant height and heads per plant in other studies. It is notable that in this study the difference of genetic and phenotypic correlation coefficients between plant height and heads per plant was not significant that it was due to partly low effect of environment on this relation (Table 4). 1000-seeds weight had the most correlation coefficient (rg=0/79, rp=0/80) with seed yield per plant.
| SYP | SW | SH | MHD | PH | HP | DM | DIF | Traits |
|---|---|---|---|---|---|---|---|---|
| -0.18ns | 0.04ns | -0.48* | -0.29ns | 0.47* | 0.23ns | 0.44* | 1 | DIF |
| 0.20ns | 0.40ns | 0.11ns | 0.37ns | 0.30ns | 0.15ns | 1 | 0.41 | DM |
| 0.10ns | 0.51** | 0.05ns | 0.16ns | 0.48* | 1 | 0.13 | 0.18 | HP |
| -0.15ns | 0.27ns | -0.05ns | -0.01ns | 1 | 0.46* | 0.30 | 0.45* | PH |
| 0.72** | 0.78** | 0.66** | 1 | -0.01 | 0.14 | 0.36 | -0.27 | MHD |
| 0.03ns | 0.28ns | 1 | 0.65** | -0.05 | 0.03 | 0.11 | -0.45* | SH |
| 0.80** | 1 | 0.06 | 0.72** | -0.15 | 0.08 | 0.20 | -0.17 | SW |
| 1 | 0.79** | 0.26 | 0.77** | 0.26 | 0.49* | 0.35 | 0.02 | SYP |
Table 4: Results of phenotypic (below diameter of table) and genetic correlation (above diameter of table) coefficients among nine traits of 20 safflower genotypes.
High correlation between 1000-seeds weight and seed yield traits reported in another study [11]. After 1000-seeds weight, main head diameter and heads per plant traits showed maximum genetic and phenotypic correlation with seed yield per plant, respectively (Table 4). Generally the results of calculating correlation coefficient in this study suggested that 1000-seeds weight, main head diameter and heads per plan traits had a more relative importance in determining the performance of safflower seeds, respectively. So, in the safflower breeding program we can use them as selection indexes to improve the seed yield.
Cluster analysis
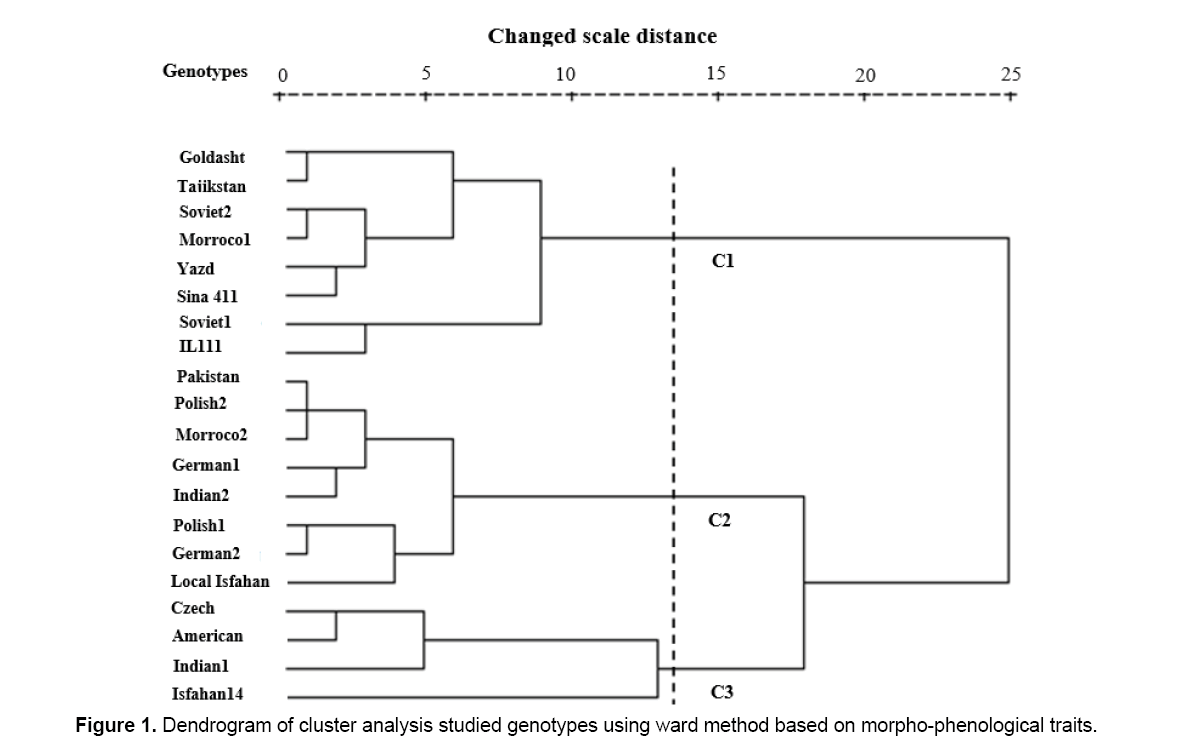
By cutting of the dendrogram resulted from cluster analysis (Ward method) in changed scale distance 14, three genotypes cluster obtained which implied genetic variation among studied genotypes (Figure 1). Results of F Beele's type statistic showed that there was a significant difference between genotypes located in these three groups for the majority of traits except three traits including days to maturity, plant height and heads per plant (Table 5).
|
Trait |
Mean square between groups |
Mean square among groups |
mean
Group1 |
Mean Group2 |
mean Gropu3 |
|---|---|---|---|---|---|
|
DIF |
** 177 | 14 | 59.81b | 62.6b | 71.25a |
|
DM |
81ns | 32.62 | 116a | 114a | 121.8a |
|
PH |
53.25ns | 91.4 | 71.07a | 70.75a | 76.68a |
|
HP |
0.91ns | 2.97 | 11.08a | 11.34a | 10.52a |
|
MHD |
12029** | 690 | 276.11a | 201.2b | 214.5b |
|
SH |
103** | 17.73 | 37.7a | 34.6ba | 30b |
|
SW |
241** | 16 | 40.36a | 29.4b | 35.8ab |
|
SYP |
74** | 9.04 | 15.6a | 9.51b | 12.6ab |
Table 5: Result of F Beele's type statistic on the groups obtained from cluster analysis.
Therefore, it was concluded that grouping was selected correctly. In another study conducted on genetic variation of safflower, genotypes located in three groups and a significant difference between genotypes for all traits except number of days to germination was reported. First group (C1) included Goldasht, Khatam Yazd, Sina 411, IL 111, Tajikistan, Soviet 1, Soviet 2 and Morocco 1. Among these genotypes, Goldasht, Khatam Yazd, Sina 411 and IL 111 belong to Iran. Most of Iranian genotypes are in this group. Genotypes from Tajikistan and former Soviet Union are in this group as well.
As regards Iran is one of the main origins of safflower and Tajikistan and former Soviet are Iran’s neighbors, it seems that these genotypes transferred from Iran to these countries by researchers who gathered germplasm in Iran. First group genotypes had higher seed yield per plant, 1000 seeds weight, seeds per head and main head diameter than other studied genotypes. Soviet-1 genotype with highest yield was located in this group. Studied genotypes of this group had least amount of days to initiate flowering trait (Table 3). Second group genotypes (C2) included Pakistan, Poland-2, Morocco-2, Germany-1, India-2, Poland-1, Germany-2 and territorial of Isfahan (Figure 1). Second group was those genotypes with maximum number of heads per plant (Table 3). Czech Republic, America, India-1 and Isfahan–14 genotypes were located in group three (C3) (Figure 1). These genotypes had least amount of seeds per head and heads per plant traits (Table 3). Also this group had the maximum amount of plant height, days to initiate flowering and days to maturity (Table 3). Isfahan–14 genotype as the most serotinal genotype of this study was located in this group. Results of genetic variation and geographical distance indicated that geographical factor is not the only factor causing genetic variation and it is not a reason of genetic far and closeness of individuals. The results showed that some of Iranian genotypes were placed in foreign genotypes groups and some of foreign genotypes located in Iranian genotypes group, so there is no conformity between genetic variation and geographical variation (Figure 1). Inconsistent between genetic variation and geographical variation reported in some other studies [5]. Locating genotypes of different ecological conditions and different countries in same cluster is probably due to their similar genetic basis that transferred to different regions of the world by researchers who gathered germplasm. The resulted of dendrogram derived from genotypes cluster analysis revealed that third group genotypes had the maximum genetic distance from the first and second group’s genotypes (Figure 1). So, applying crosses programs between genotypes of these clusters will provide higher heterosis and genetic variation. It was suggested that more genetic distance of parents lead to more heterosis of offspring [5]. In the first group genotypes, the more seed yield per plant is due to the more main head diameter, 1000-seeds weight and seeds per head in this group. Soviet-1 and IL–111 genotypes are located in one group had the greatest seed yield per plant, good productive potency, respectively. Therefore, they can be used in safflower breeding programs. Generally, the results of this study showed that studied genotypes had a great genetic variation in view of seed yield and its main component and as well as some of these genotypes such as Soviet-1 and IL–111 containing high productive potency or other desirable traits. So, it can be used in breeding programs and produce improved varieties with desirable traits. Correlation coefficients analysis revealed that traits of 1000-seeds weight, main head diameter and heads per plant had a more relative importance in determining the performance of safflower seeds, respectively. Therefore, we can use them as selection indexes to improve the seed yield in the safflower breeding program. Results of evaluation of genetic variation and far and near geographic distances of genotypes showed that generally geographic distance is not the reason for genetics farness and closeness of individuals. Also, results revealed that some of Iranian genotypes were placed in groups of foreign genotypes and some foreign genotypes were located into groups of Iranian genotypes as well. Situation of genotypes related to different ecological conditions and different countries in the same cluster could be due to existence of their identical genetic basis and as well as the exchange of plant material across these origins. Generally, since morpho-phenological traits are affected by environmental factor than molecular markers, therefore, we suggested that molecular markers are used for assessment of phylogenic relationships in future investigation.
References
- Bowles V (2010) Relationship and introgression within Carthamus (Asteraceae), with an emphasis on safflower (Carthamustinctorious L.). A Thesis of M.S, University of Alberta.
- Jaradat A, Shahid M. (2006). Pattern of phenotypic variation in a germplasm collection of Carthamustinctorious L. From the Middle East. Genetic Resources and Crop Evolution.53:225-244.
- Dwivedi SL, Upadhyaya HD, Hegde DM. (2005). Development of core collection using geographic information and morphological descriptors in safflower (Carthamustinctorius L.) germplasm. Directorate of Oilseed Research, Rajandra Nagar, Hyderabad 500 030, India. Genetic Resource and Crop Evolution.52: 821-830.
- Bahmankar M, NabatiAhmadi D, Dehdari M. (2014). Correlation, multiple regression and path analysis for some yield-related traits in safflower. Journal of Biodiversity and Environmental Sciences.4: 111-118.
- Bagheri A, YazdiSamadi B, Taeb M. (2001).A study of genetic diversity in landrace populations of safflower in Iran. Iranian JournalofAgricultural Science.32:456-447.
- Golkar P.(2012).Assessment of genetic diversityof cultivatedsafflower genotypes(Carthamustinctorius L.)usingmorphological traits,17thNational andFifth International ConferenceofBiology, ShahidBahonarUniversity, Kerman, Iran.
- ShahbaziDur BashS, AlizadehDizajiKH, SadeghzadehB, et al. (2011).Evaluation of genetic diversity in safflower landraces using agro-morphological characters and RAPD markers.Iranian Journal of Field CropScience.42:231-221.
- Rafiei F, Saeedi G. (2005). Agenetic variation for different agronomic traits in isolated lines of safflower (Carthamustinctorious l.) from Iranian local populations and foreign genotypes. Journal of Science and Technology of Agriculture and Natural Resources. 9:91-106.
- Elfadl E, Reinbrecht C,Claupein W. (2009). Evaluation of phenotypic variation in a worldwide germplasm collection of safflower (Carthamustinctorius L.) grown under organic farming conditions in Germany. Genetic Resource and Crop Evolution.57: 155–170.
- Rafiei F,Saeedi G (2005) B-Genotypic and phenotypic relationships among agronomic traits and yield components in safflower (Carthamustinctorious L.). Agriculture Scientific Journal.28:137-149.
- Acharya S, Dhaduk LK, Maliwal GL. (1994). Path analysis in safflower under conserved moisture conditions. Gujarat-Agricultural University Results.20: 154-157.
- Safavi SM, Pourdad SS, Safavi SA. (2012). Assessment of genetic diversity in safflower (Carthamustinctorius L.) genotypes using agro-morphological traits. Annals of Biological Research.3:2428-2432.
- Saeedi G, Tofi H,Mirlohi. (2004). Genetic variation and relationships among characters in some safflower land races of Iran. Journal of Agricultural Sciences and Natural Resources.11: 116-107.
- Falconer DS, Mackay TFC.(1996).Introduction to Quantitative Genetics, Ed4, Longman, Harlow, UK.
- FarshadfarA(1996)Methodologyof plant breeding, University of KermanshahandTaghebostanPress.616.
- Dehdari A, Rezaei A, Mobli M.(2001).Morphological and agronomic characters of landrace varieties of onion (Allium cepa L.) and their classification. Journal of Science and Technology of Agriculture and Natural Resources.5: 123-109.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences