Computational Drug Discovery of Potential Aldose Reductase Inhibitors Using in silicoStudies
Arumugam Madeswaran, Muthuswamy Umamaheswari, Kuppusamy Asokkumar, Thirumalaisamy Sivashanmugam, Varadharajan Subhadradevi, Puliyath Jagannath
Department of Pharmacology, College of Pharmacy, Sri Ramakrishna Institute of Paramedical Sciences, Coimbatore, Tamil Nadu, India
- Corresponding Author:
- Tel: +91 9787685014
E-mail: madeswaran2@gmail.com
Abstract
The objective of the current study was to investigate the aldose reductase inhibitory activity of flavonoids using in silico docking studies. In this perspective, flavonoids like 4-methyl esculatin, silbinin, taxifolin, and wogonin were selected. Epalrestat, a known aldose reductase inhibitor was used as the standard. In silico docking studies were carried out using AutoDock 4.2, based on the Lamarckian genetic algorithm principle. The results showed that, the selected flavonoids showed binding energy ranging between -10.60 kcal/mol to -7.17 kcal/mol when compared with that of the standard (-8.73 kcal/mol). Inhibition constant (16.94 nM to 5.51 μM) and intermolecular energy (-12.99 kcal/mol to -8.37 kcal/mol) of the flavonoids also coincide with the binding energy. In the selected flavonoids, silbinin and taxifolin were contributed better aldose reductase inhibitory activity than the standard because of its structural properties. These molecular docking analyses could lead to the further development of potent aldose reductase inhibitors for the treatment of diabetes.
Keywords
Binding energy, flavonoids, diabetes, silbinin, ligands.
1. Introduction
Molecular docking is a frequently used tool in computer-aided structure-based rational drug design. It evaluates how small molecules called ligands (flavonoids) and the target macromolecule (aldose reductase enzyme) fit together [1,2]. Auto Dock Tools (ADT) is a program package of automated docking tools and designed to predict how small molecules bind to a target protein of known 3D-structure. Besides generating binding energies in these docking studies, the position of the ligand in the enzyme binding site can be visualized [3]. It can be useful for the further development of potential drug candidates and also for the understanding of the binding nature of the ligands with the target.
Diabetes mellitus is a metabolic disorder characterized by hyperglycemia resulting from defects in insulin action, insulin secretion or both. Type 1 diabetes is caused by a deficiency of β-pancreatic cells insulin secretion. Type 2 diabetes is associated with obesity and is characterized by an initial phase progressive insulin resistance, with ensuing reduction in the ability of pancreatic hormone to promote peripheral glucose disposal and to decreases hepatic glucose output [4,5].
Aldose reductase (ALR2; EC: 1.1.1.21) belongs to aldo-keto reductases super family. It is the first rate limiting enzyme in polyol pathway and reduces glucose to sorbitol by utilizing NADPH as a cofactor. sorbitol dehydrogenase is the enzyme responsible for the conversion of Sorbitol into fructose [6,7]. The polyol pathway represents a minor route of glucose utilization, accounting for less than 3% of glucose consumption. However, in the presence of high glucose, the activity of this pathway is increased and could represent up to 30% of total glucose consumption [8]. Abnormal activation of the polyol pathway during diabetes leads to accumulation of osmotically active sorbitol leading to osmotic as well as oxidative stress, resulting in tissue injury [9,10]. Evidence for the involvement of ALR2 in diabetic neuropathy, retinopathy, nephropathy and cataract emerged from several independent studies [11].
Flavonoids and their related compounds are low molecular weight substances, which are a group of natural products which exhibits various biological and pharmacological activities like antibacterial, antiviral, antioxidant, anti inflammatory, anti allergic, hepatoprotective, antithrombotic, antiviral and antimutagenic effects and inhibition of several enzymes have also been investigated [12-15]. The objective of the present work is to study the in silico aldose reductase inhibitory activity of commercially available flavonoids.
2. Materials and Methods
Softwares Required.
Python 2.7 - language was downloaded from www.python.com, Cygwin (a data storage) c:\program and Python 2.5 were simultaneously downloaded from www.cygwin.com, Molecular graphics laboratory (MGL) tools and AutoDock4.2 was downloaded from www.scripps.edu, Discovery studio visualizer 2.5.5 was downloaded from www.accelerys.com, Molecular orbital package (MOPAC), ChemSketch was downloaded from www.acdlabs.com, Online smiles translation was carried out using cactus.nci.nih.gov/translate/.
Docking Methodology
Lamarckian genetic algorithm (LGA) for ligand conformational searching is used for the docking, which is a hybrid of genetic algorithm and local search algorithm. This algorithm first builds a population of individuals, each being a different random conformation of the docked molecule. Each individual is then mutated to acquire a slightly different translation and rotation and the local search algorithm then performs energy minimizations on a user-specified proportion of the individuals. The individuals with the low resulting energy are transferred to the next generation and the process is then repeated. The algorithm is called Lamarckian genetic algorithm because every new generation of individuals is allowed to inherit the local search adaptations of their parents [16]. An extended PDB format, termed as PDBQT file was used for coordinate files which includes atomic partial charges. AutoDock Tools was used for generating PDBQT files from traditional PDB files [17]. Crystal structure of aldose reductase enzyme was downloaded from the Brookhaeven protein data bank (Figure 1).
Even though there are thousands of structures of aldose reductase enzymes are present in the database but the 3EL3 were selected for the present study due to its similarity with the human enzyme. Albaflavenone synthase (CYP170A1) is monooxygenase catalyzing the final two steps in the biosynthesis of this antibiotic in the soil bacterium, Streptomyces coelicolor A3. Interestingly, CYP170A1 shows no stereo selection forming equal amounts of two albaflavenol epimers, each ofwhich is oxidized in turn to albaflavenone. To explore the structural basis ofthe reaction mechanism,we have studied the crystal structures of both ligand-free CYP170A1 (2.6 A) and complex ofendogenous substrate (epi-isozizaene) with CYP170A1 (3.3 A). The structure of the complex suggests that the proximal epi-isozizaene molecules may bind to the heme iron in two orientations.
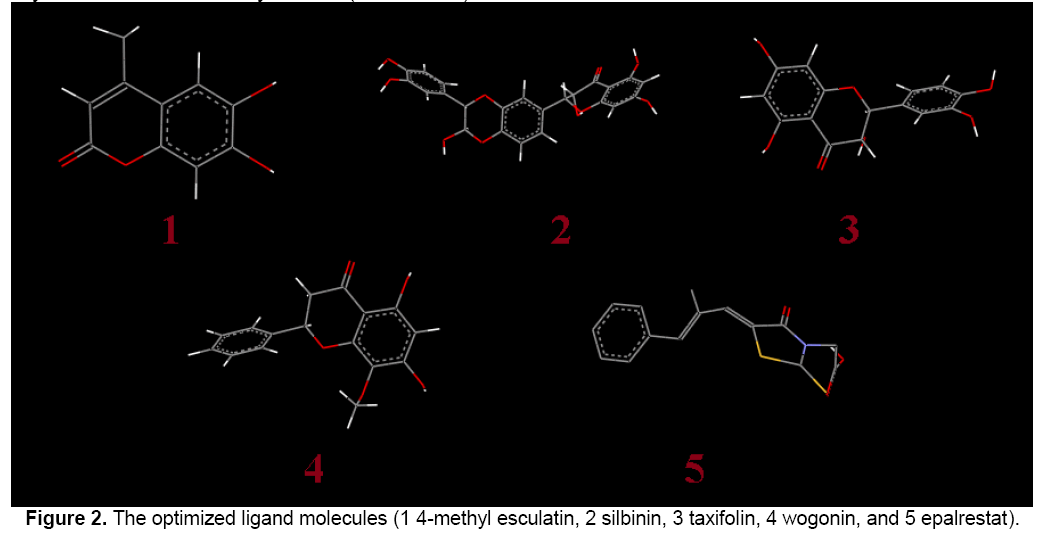
In Figure 2, the flavonoid ligands like 4-methyl esculatin, silbinin, taxifolin, wogonin and epalrestat were built using ChemSketch and optimized using “Prepare Ligands” in the AutoDock 4.2 for docking studies. The optimized ligand molecules were docked into refined aldose reductase model using “LigandFit” in the AutoDock 4.2 [18].
Lead optimization of the selected compounds was done by computation of druglikeness properties. The druglikeness scores of the compounds were evaluated with the help of Lipinski’s rule. The various parameters of the ligands like molecular formula, molecular weight, aromatic carbons, rotatable bonds and no. of torsions were tabulated in Table 1.
The enzyme was refined using various steps with the help of Accelrys studio viewer. The water molecules and heavy metal atoms have been removed from the enzyme and then hydrogen and Kollman charges has been added to the enzyme. Then the pdb format of the enzyme converted into suitable pdbqt format for the docking.
The preparation of the target protein 1EL3 with the AutoDock Tools software involved adding all hydrogen atoms to the macromolecule, which is a step necessary for correct calculation of partial atomic charges. Gasteiger charges are calculated for each atom of the macromolecule in AutoDock 4.2 instead of Kollman charges which were used in the previous versions of the program. Three-dimensional affinity grids of size 277 × 277 × 277 Å with 0.6 Å spacing were centered on the geometric center of the target protein and were calculated for each of the following atom types: HD, C, A, N, OA, and SA, representing all possible atom types in a protein. Additionally, an electrostatic map and a desolvation map were also calculated [18]. The dimensional affinity grid was fixed based upon the necessity to run the AutoGrid file.
Rapid energy evaluation was achieved by precalculating atomic affinity potentials for each atom in the ligand molecule. In the AutoGrid procedure, the target enzyme was embedded on a three dimensional grid point [17]. The energy of interaction of each atom in the ligand was encountered.
The selected important docking parameters for the LGA as follows: population size of 150 individuals, 2.5 million energy evaluations, maximum of 27000 generations, number of top individuals to automatically survive to next generation of 1, mutation rate of 0.02, crossover rate of 0.8, 10 docking runs, and random initial positions and conformations. The probability of performing local search on an individual in the population was set to 0.06.
AutoDock was run several times to get various rigid docked conformations, and used to analyze the predicted docking energy. The binding sites for these molecules were selected based on the ligand-binding pocket of the templates. AutoDock Tools provide various methods to analyze the results of docking simulations such as, conformational similarity, visualizing the binding site and its energy and other parameters like intermolecular energy and inhibition constant. For each ligand, ten best poses were generated and scored using AutoDock 4.2 scoring functions [17].
3. Results
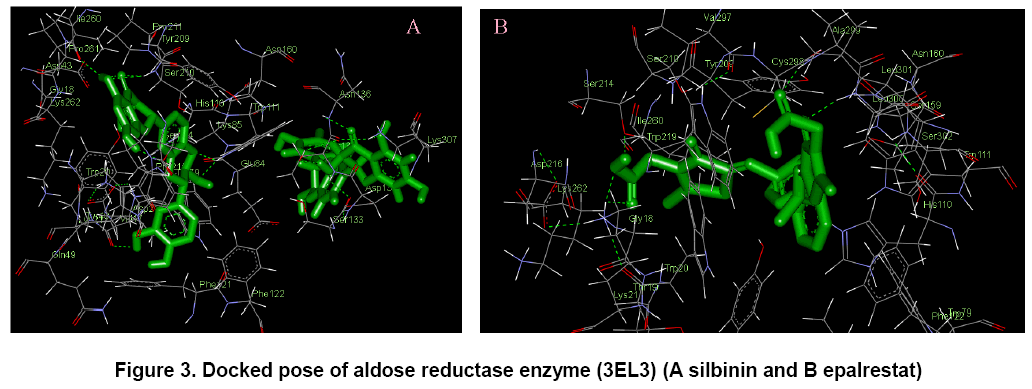
The docking poses were ranked according to their docking scores and both the ranked list of docked ligands based on their binding energy with the enzyme and their corresponding binding poses [18]. In Figure 3, docked pose of aldose reductase enzyme with the ligands silbinin and epalrestat clearly demonstrated the binding positions of the ligand with the enzyme.
Binding energy of the individual compounds were calculated by using the following formula,
Binding energy = A+B+C-D
where, A denotes final intermolecular energy + van der Waals energy (vdW) + hydrogen bonds + desolvation energy + electrostatic energy (kcal/mol), B denotes final total internal energy (kcal/mol), C denotes torsional free energy (kcal/mol), D denotes unbound system’s energy (kcal/mol).
As shown in Table 2, flavonoids showed binding energy ranging between -10.60 kcal/mol to -7.17 kcal/mol. All the selected flavonoids had showed better and consistent binding energy when compared to standard epalrestat (-8.73 kcal/mol). This proves that flavonoids consist of potential aldose reductase inhibitory binding sites similar to that of the standard.

In addition, two other parameters like inhibition constant (Ki) and intermolecular energy were also determined. Inhibition constant is directly proportional to binding energy. As shown in Table 3, flavonoids showed inhibition constant ranging from 16.94 nM to 5.51 μM. All the selected compounds had lesser inhibition constant when compared to the standard (397.18 nM). Thus, the aldose reductase inhibitory activity of the flavonoids were compared with the standard.
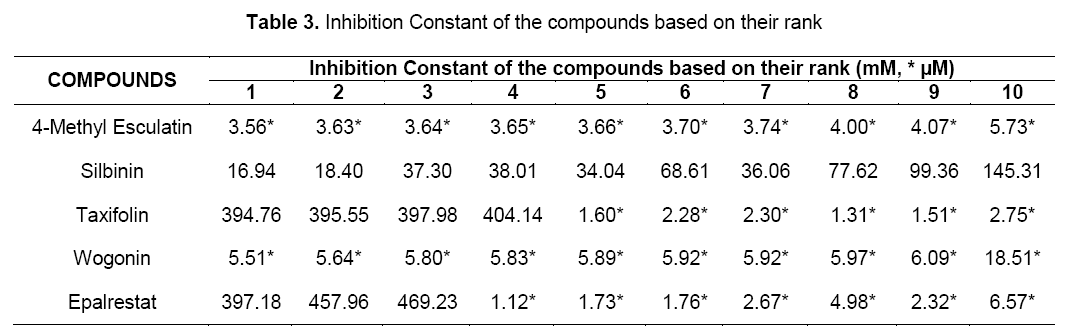
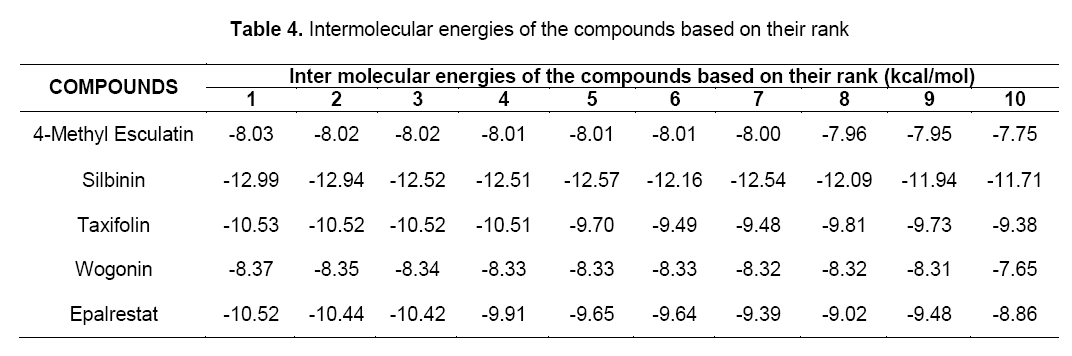
Intermolecular energy is also directly proportional to binding energy. As shown in Table 4, flavonoids showed intermolecular energy ranging between -12.99 kcal/mol to -8.37 kcal/mol which was lesser when compared to the standard (-10.52 kcal/mol). We found a decrease in intermolecular energy of all the selected compounds with a simultaneous decrease in the binding energy. This result further proved the aldose reductase inhibitory activity of all the selected flavonoids.
4. Discussions and Conclusions
Analysis of the receptor/ligand complex models generated after successful docking of the flavonoids was based on the parameters such as hydrogen bond interactions, п – п interactions, binding energy, RMSD of active site residues and orientation of the docked compound within the active site [19].
In silico docking study, was carried out to identify the inhibiting potential of selected flavonoids against aldose reductase enzyme. In this study 4 different flavonoids were selected for the in silico docking studies. Lead optimization of the selected compounds was done by computation of druglikeness properties. The druglikeness scores of the compounds were evaluated with the help of Lipinski’s rule. The docking studies were performed by the use of AutoDock4.2. In the docking studies, if a compound shows lesser binding energy compared to the standard it proves that higher possibility of the compound has higher activity.
The potential binding sites of the silbinin (Fig. 3 A) was found that, Gly 18, Trp 20, Lys 21, Gln 26, Val 27, Asp 43, Tyr 48, Gln 49, Glu 84, Lys 85, His 110, Trp 111, Phe 121, Phe 122, Ser 133, Asn 160, Tyr 209, Ser 210, Pro 211, Ser 214, Pro 215, Ile 260, Pro 261, Lys 262, Lys 307. The binding sites of the epalrestat (Fig. 3 B) was found to be Gly 18, Thr 19, Trp 20, Lys 21, Tyr 48, Gln 49, Glu 84, His 110, Trp 111, Phe 121, Phe 122, Ser 133, Asn 160, Tyr 209, Ser 210, Pro 211, Ser 214, Pro 215, Asp 216, Trp 219, Ile 260, Pro 261, Lys 262, Cys 298, Val 297, Ala 299, Leu 301, Ser 302, Lys 307. This proves that the effective binding sites are present in the selected flavonoid silbinin when compared with the standard epalrestat. It proves that the ability of inhibiting the aldose reductase enzyme by the selected flavonoid.
Based on the current docking studies, the aldose reductase inhibitory activity of the selected compounds was found to be decreased in the order of silbinin, taxifolin, epalrestat, 4-methyl esculatin, wogonin. On the basis of the above study, all the selected flavonoids showed better aldose reductase inhibitory activity than the standard. Among the selected flavonoids, silbinin and taxifolin showed excellent binding interactions with aldose reductase enzyme than the standard. This may be attributed due to the differences in the position of the functional groups in the compounds.
These results clearly indicate that the flavonoids especially, silbinin and taxifolin showed excellent binding interactions with aldose reductase enzyme than the standard. This in silico studies is actually an added advantage to screen the aldose reductase enzyme inhibition. Further investigations on the above compounds and in vivo studies are necessary to develop potential chemical entities for the prevention and treatment of diabetes.
References
- Norgan A.P., Coffman P.K., Kocher J.P.A., et al. (2011) Multilevel Parallelization of AutoDock 4.2. J Cheminform, 3: 12-15.
- Seeliger D., de Grootligand B.L. (2010) Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des, 24: 417-422.
- Zhang S., Kumar K., Jiang X., et al. (2008) DOVIS: An implementation for high throughput virtual screening using Autodock. BMC Bioinform, 9: 126-128.
- Vianna R., Brault A., Martineau L.C., et al. (2011) In vivo anti-diabetic activity of the ethanolic crude extract of Sorbus decora c.k. schneid. (rosacea): A medicinal plant used by canadian james bay cree nations to treat symptoms related to diabetes. Inventi Impact Ethno Pharmacol, 3: 10-15.
- Lamba H.S., Bhargava C.S., Thakur M., et al. (2011) αglucosidase and aldose reductase inhibitory activity in vitro and antidiabetic activity in vivo of tribulus terrestris L. (dunal). Int J Pharm Pharmaceut Sci, 3: 270-272.
- Hwang C.Y., Shaw S., Kaneko M., et al. (2005) Aldose reductase pathway mediates JAK-STAT signaling: a novel axis in myocardial ischemic injury. J Fed Am Soc Exp Biol, 19: 795-797.
- Ravindranath T.M., Mong P.Y., Ananthakrishnan R., et al. (2009) Novel Role for Aldose Reductase in Mediating Acute Inflammatory Responses in the Lung. J Immunol, 183: 8128-8137.
- Yadav U.C.S., Naura A.S., Aguirre L.A., et al. (2009) Aldose reductase inhibition suppresses the expression of Th2 cytokines and airway inflammation in ovalbumin-induced asthma in mice. J Immunol, 183: 4723-4732.
- Dong Y., Yang J., Ren X., et al. (2005) New Aldose Reductase Inhibitors N99-596 A and B from Streptomyces. J Antibio, 58: 737-739.
- Saraswat M., Muthenna P., Suryanarayana P., et al. (2008) Dietary sources of aldose reductase inhibitors: prospects for alleviating diabetic complications. Asia Pacific J Clin Nut, 17: 558-565.
- Guzman A., Guerrero O.R. (2005) Inhibition of aldose reductase by herbs extracts and natural substances and their role in prevention of cataracts. Revista Cubana de Plantas Medicinales, 10: 3-4.
- Cushnie T.P., Lamb A.J. (2011) Recent advances in understanding the antibacterial properties of flavonoids. Int J Antimicrob Agents, 38: 99-107.
- Liu H., Wang Z., Qiao Y., et al. (2007) Flavonoids with Aldose Reductase Inhibiting Activity: Pharmacophore Modeling and Implications for Mechanism. Acta Physico-Chimica Sinica, 23: 1059-1064.
- Gonzalez R., Ballester I. (2011) Effects of Flavonoids and other Polyphenols on Inflammation. Crit Rev Food Sci Nut, 51: 331-362.
- Nishiumi S., Miyamoto S., Kawabata K., et al. (2011) Dietary flavonoids as cancer-preventive and therapeutic biofactors. Front Biosci, 3: 1332-1362.
- Madeswaran A., Umamaheswari M., Asokkumar K., et al. (2011) Docking studies: In silico lipoxygenase inhibitory activity of some commercially available flavonoids. Bangladesh J Pharmacol, 6: 133-138.
- Umamaheswari M., Madeswaran A., Asokkumar K., et al. (2012) Docking studies: Search for possible phytoconstituents for the treatment of gout. Int J Biol Pharm Res, 3: 6-11.
- Madeswaran A., Umamaheswari M., Asokkumar K., et al. (2012) In Silico docking studies of lipoxygenase inhibitory activity of commercially available flavonoids. Orient Pharm Exp Med, 12: 157-61.
- Azam F., Prasad M.V.V., Thangavel N. (2011) Molecular docking studies of 1- (substituted phenyl) -3- (naphtha [1,2 –d] thiazol -2-yl) urea/thiourea derivatives with human adenosine A2A receptor. Bioinformation, 6: 330-334.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences