Candida parapsilosis Complex Distribution, Behavior and Response Profile to Antifungal Agents in a Collection of Blood Cultures from Argentinean Patients with Candidemia
Rodríguez L, Rosa AC, Mayo S, Sellart G, Magariños F, Santillán HD, Bertone A, Jewtuchowicz V
Rodríguez L1,2,*, Rosa AC3, Mayo S4, Sellart G4, Magariños F5, Santillán HD5, Bertone A1, Jewtuchowicz V1,6
1Buenos Aires University, School of Medicine, Center for Micology, Institute of Microbiology, Parasitology and Immunology (IMPaM), UBA-CONICET, Buenos Aires, Argentina
2Department of Semiology and Clinical Diagnosis, School of Dentistry, Cuenca University, Ecuador
3Buenos Aires University, School of Dentistry, Department of Microbiology and Immunology. Buenos Aires, Argentina
4Hospital Luisa C de Gandulfo, Infectology Ward, Buenos Aires Province, Argentina
5 Hospital Luisa C de Gandulfo, Oncology Ward, Buenos Aires Province, Argentina;
6Hospital Luisa C de Gandulfo, Laboratory Service, Buenos Aires Province, Argentina.
Received date: March 23, 2017; Accepted date: April 07, 2017; Published date: April 14, 2017
Citation: Rodríguez L, Rosa AC, Mayo S, et al. Candida parapsilosis Complex Distribution, Behavior and Response Profile to Antifungal Agents in a Collection of Blood Cultures from Argentinean Patients with Candidemia. Electronic J Biol, 13:2
Abstract
Candida parapsilosis is a complex comprising three yeast species which can be distinguished genetically. These yeasts have emerged over the past decade as major nosocomial pathogens, with the species C. parapsilosis sensu stricto being the second most frequently isolated yeast after C. albicans from blood samples of patients with candidemia. In Argentina there are few data available on the epidemiology of this complex in samples from patients with invasive disease. Our aim was to analyze the distribution, relationship with immune status and response to antifungal agents commonly used in clinical practice, of the species in the C. parapsilosis complex from a collection of blood cultures from Argentinean patients diagnosed with candidemia. A basic, retrospective, cross-sectional study was designed for molecular analysis of 25 blood cultures by endpoint PCR with specific primers from the ITS1-5.8SrRNA-ITS2 region. Minimum Inhibitory Concentration (MIC) was determined for each antifungal agent on the study strains using Vitek2 automated method. In Argentina C. parapsilosis sensu stricto is the predominant species in samples from patients with invasive disease, being more likely to be recovered from clearly immunocompromised patients. Flucytosine does not seem to be a good choice for treating invasive infections by this yeast.
Keywords
Candida parapsilosis complex; Blood culture; Candidemia; Vitek2.
1. Introduction
Candida species have emerged as major pathogens over the past two decades and Invasive Candidiasis (IC) is one of the most common causes of invasive infectious nosocomial diseases [1,2]. Although Candida albicans is still the predominant etiological species, >50% of candidemia and IC cases may be the result of Non-Candida albicans Candida (NCAC) species and within this group, Candida parapsilosis is the second or third most commonly isolated yeast from blood cultures in Latin America, Canada Europe and Asia [1,2]. In Argentina, an important study published in 2004 used standard techniques to characterize 1006 isolates from a wide range of clinical samples taken during 1999-2001, reporting 54.9% prevalence of NCAC species, with C. parapsilosis being the species most frequently found in blood cultures and onyxis, having higher frequencies than C. albicans at both those anatomical sites. C. parapsilosis is responsible for a wide range of clinical manifestations, usually occurring in patients who are catheterized, patients with deteriorated immune system, neutropenia, burns, pre-term neonates or patients in intensive care units [2,3].
Tavanti et al. [3] conducted studies based on analysis of the ITS1 sequence in C. parapsilosis isolates from groups I, II and III, as they were considered at that time, and compared them to another 15 species of Candida and Sacharomyces cerevisiae. They found that Candida parapsilosis is a complex comprised of 3 different species which are phenotypically indistinguishable. The complex was therefore restructured, with C. parapsilosis sensu stricto being assigned to C. parapsilosis group I; Candida orthopsilosis to C. parapsilosis group II; and C. metapsilosis to group III isolates. C. orthopsilosis and C. metapsilosis were the most similar to each other [3].
The literature reports that of the three species, C. parapsilosis sensu stricto is the most frequently isolated worldwide from different human ecological niches in conditions of both health and disease, and especially in immunocompromised patients [4-6]. The pattern of distribution of C. orthopsilosis and C. metapsilosis is more heterogeneous, varying according to geographic region, clinical service, anatomical site and host immune status [1,5].
In Argentina, few papers analyze the distribution and behavior of the different species in this complex in samples from patients with invasive disease. The current study therefore looks at the prevalence of isolates of the three species in the parapsilosis complex, their relationship to host immune status and their response profile to antifungal agents, based on a collection of blood cultures from Argentine patients with candidemia.
2. Materials and Methods
A basic, retrospective, cross-sectional study was designed using a sample of 25 blood cultures characterized phenotypically by conventional methods as Candida parapsilosis complex. The samples were obtained from a population of 101 cryopreserved Candida parapsilosis isolates from different human ecological niches: urine, blood, skin, nail and oral cavity, from the yeast collection at the Center for Mycology, School of Medicine, Buenos Aires University, Argentina and the Hospital Luisa C. de Gandulfo, Buenos Aires Province, Argentina.
The clinical histories of the patients in whom the isolates were found were available. The following ATCC reference strains were used: C. parapsilosis (ATCC 22019), C. orthopsilosis (ATCC 96139) and C. metapsilosis (ATCC 96143). They were subjected to the same procedures as the clinical isolates.
For the in vitro susceptibility test, minimum inhibitory concentration (MIC) values were obtained using the automated Vitek2 method (bioMérieux)-Card AST-YS07, which some studies report has a correspondence greater than 90% with the reference method (broth microdilution) proposed in CLSI document M27-A3 (2008) [7,8]. Said system was used to evaluate the response of 25 clinical isolates to the following antifungal agents: Fluconazol (FZ), Voriconazol (VZ), Caspofungin (CAS), Micafungin (MICA), Amphotericin B (AB) and Flucytosine (FC). Readings were interpreted according to the 2012 update on species-specific Clinical Breakpoints (CBPs) and epidemiological cutoff value (ECV), which were later included in the 2012 CLSI M27-S4 document [12]. The following Candida strains were used as quality controls for the study: Candida krusei ATCC 6258, C. parapsilosis ATCC 22019 and C. albicans ATCC 9002.
The following variables were analyzed: C. parapsilosis complex, immune status and antifungal activity. The associations between C. parapsilosis complex and immune status and between C. parapsilosis complex and antifungal activity were studied.
2.1 Distribution of parapsilosis complex species in 25 blood cultures from patients with candidemia
The cryopreserved isolates were firstly identified based on color when developing in chromogenic medium, micro-morphology in 1% milk agar-Tween 80, and carbohydrate assimilation profiles according to commercial API ID 32D and Vitek2 (BioMérieux, Francia) systems [9,10].
The molecular characterization of each of the isolates required the following steps: (1) Obtaining DNA. DNA was obtained using the zymolase technique to break down cell walls, and purified using the commercial system (Qiagen) based on separating using columns (QIAamp DNA Mini Kit), following the manufacturer’s instructions. The DNA obtained was preserved at -20ºC. (2) Molecular identification of the species in the complex. Molecular typing was done by end-point PCR using specific primers derived from unique sequences contained in the internal transcriptional spacer 1 (ITS 1)-5.8 rRNA-(ITS2) of the fungal ribosomal DNA; which enable a sequence specific to the species C. parapsilosis sensu stricto, C. orthopsilosis and C. methapsilosis to be retrieved separately (Table 1) [11]. (3) Amplification characteristics and conditions. The protocol proposed by Asadzadeh et al. [11] was used to amplify the region contained in the internal transcriptional spacer ITS1-5.8rRNA-ITS2 with the set of specific primers. Said protocol applies an optimized technique which was validated using an alternative PCR-RFLP method and the reference method (Sanger sequencing). Table 2 shows final concentrations of PCR reaction.
Cycling conditions were: one 5 min cycle at 95ºC, followed by 30 cycles, each with 3 stages of 94ºC: (1 min); 63ºC (45 s); 72ºC (1 min) and a final 10 min cycle at 72ºC.
The results of amplification with specific primers were validated with Sanger sequencing, by performing end-point PCR using the pan-fungal primers ITS 1 and ITS 4 for amplification followed by sequencing of the ITS1-ITS4 region of ribosomal RNA gene 28S, as described by White et al [25]. Table 3 shows the protocol used for PCR amplification of the ITS region.
| Primer | Target Gene | Direction | Species Specificity | Sequence | Amplicon Size |
|---|---|---|---|---|---|
| CPAF | ITS 1 | Forward | C. parapsilosis | TTTGCTTTGGTAGGCCTTCTA | 379 pb |
| CPAR | ITS 2 | Reverse | GAGGTCGAATTTGGAAGAAGT | ||
| CORF | ITS 1 | Forward | C. orthopsilosis | TTTGGTGGCCCACGGCCT | 367 pb |
| CORR | ITS 2 | Reverse | TGAGGTCGAATTTGGAAGAATT | ||
| CMEF | ITS 1 | Forward | C. methapsilosis | TTTGGTGGGCCCACGGCT | 374 pb |
| CMER | ITS 2 | Reverse | GAGGTCGAATTTGGAAGAATGT |
Table 1: Primers used for rapid identification at species level of the C. parapsilosis complex.
| Reagents | Final concentration |
|---|---|
| Water | |
| PCR Buffer | 1X |
| Cl2Mg | 3 mM |
| dNTPs | 0.1 mM |
| First complex C. parapsilosis (F) |
10 pmol |
| First complex C. parapsilosis (R) |
10 pmol |
| Taq DNA pol | 1.25 |
| DNA or suspension | 10 ng/ml |
Table 2: PCR protocol: Reagent concentrations.
| Components | Final concentration |
|---|---|
| Water | |
| PCR Buffer | 1X |
| Cl2Mg | 3 mM |
| dNTPs | 0.2 mM |
| Primer ITS1 | 0.4 mm |
| Primer ITS 4 | 0.4 mm |
| Taq-pol | 1.25 |
| DNA | 10 ng/ml |
Table 3: PCR protocol for amplification of the ITS region: Concentrations of reagents.
The primer sequence to hybridize with the ITS region is: ITS 1 forward: TCCGTAGGTGAACCTGCGG; and ITS 4 reverse: TCTTTTCCTCCGCTTATTGATATG. These primers amplified a 536 pb fragment of a region of the fungal ribosomal DNA. The PCR cycles were performed in the thermal cycler Mini CyclerTM, MJ Research INC, with the following protocol: one 5 min cycle at 95ºC; followed by 30 cycles, each with 3 stages of: 20 s at 95ºC; 15 s at 55ºC; and 65 s at 72ºC and one final 5 min cycle at 72ºC.
The amplified fragments were purified using the commercial device QIAquick PCR purification Kit (Qiagen) and sequenced using an ABI Prism 3730xl DNA analyzer (Applied Biosystems, BsAs-Argentina) with the ITS1 primer.
The sequences obtained were analyzed with the BLAST (Basic Local Alignment Search Tool) sequence comparison algorithm (https://www.ncbi.nlm.nih.gov/BLAST).
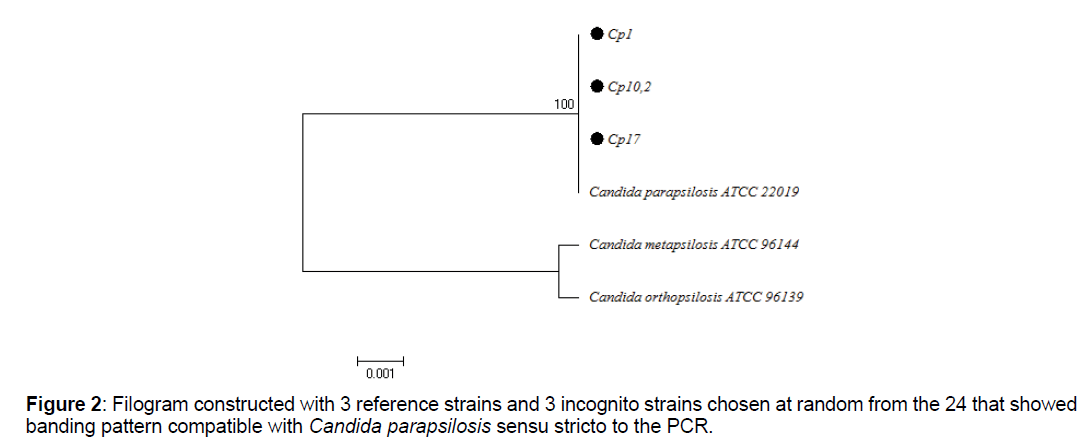
For the phylogenetic analysis, the BIOEDIT program was used for editing sequence alignment and the MEGA 6 program for multiple alignments of sequences and phylogenetic analysis, with the Neighbor Joining algorithm. The tree was constructed with the ATCC reference sequences for C. parapsilosis ATCC 22019, C. metapsilosis ATCC 96144and C. orthopsilosis ATCC 96139, in addition to the sequences selected at random from the total that were positive for PCR with specific primers.
2.2 Determination of antifungal activity of six drugs commonly used in clinical practice
For each clinical isolate, in vitro susceptibility was evaluated using the automated Vitek2 (Biomeriux) system against 6 antifungal agents (FZ, VZ, CAS, MICA, AB, FC) commonly used in clinical practice for patients with mycosis. The following reference strains were used as quality controls: Candida albicans ATCC 90028, Candida parapsilosis ATCC 22019 and C. krusei ATCC 6258.
Strains were recorded as Susceptible (S), Susceptible Dose-Dependent (SDD), Intermediate (SI) or Resistant (R), by using the species-specific clinical breakpoints reestablished by the CLSI Subcommittee led by Pfaller and Diekema and published in the Journal of Medical Microbiology-2012 [12].
Strain behavior with relation to host immune status was ascertained from data available in the clinical histories plus statistical analysis to establish presence or absence of association among variables.
3. Statistical Analysis
Data were processed on Microsoft Excel 2010 and Stadistit 7.0 software for quantitative and qualitative analysis using a 95% Confidence Interval (CI) for each, considering a p value less than alpha error (alpha error=0.05).
Frequency tables and bivariate bar graphs were used to represent variables. Association between variables was estimated using contingency tables, Chi-square test and Prevalence Ratio (PR) to determine strength of association.
4. Results
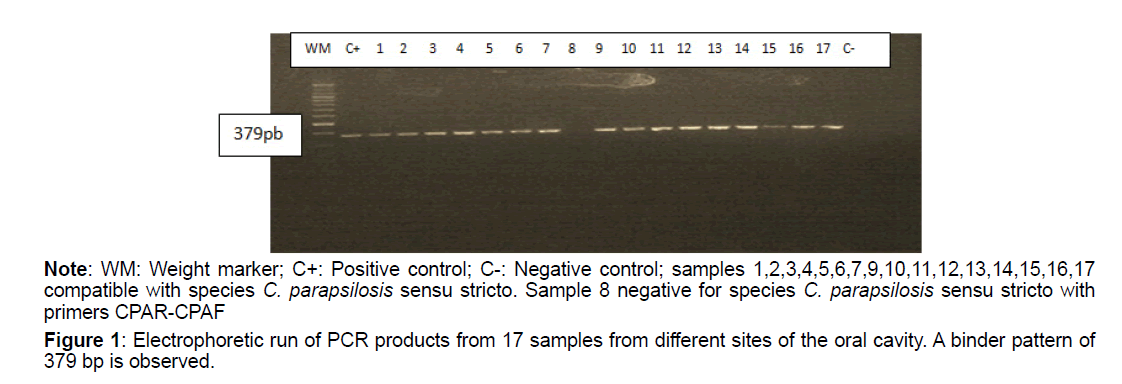
Of the total isolates subject to molecular analysis, 24 out of 25 (96%) were reconfirmed as C. parapsilosis sensu stricto (Table 4), using the end-point PCR with the specific primer pair CPAR-CPAF (Figure 1), obtaining 379pb amplicons (Figures 1-3). This band pattern is compatible with the one reported by Asadzadeh et al [11]. Only one strain (4%) identified phenotypically as C. parapsilosis did not show bands in the specific PCR and its identity was confirmed 100% as Candida albicans by sequencing. PCR results were confirmed with the phylogenetic analysis performed with the sequences obtained by Sanger sequencing, using the Bioedit and Mega 6 programs (Figure 2).
| Species | Absolute Frequency | Relative Frequency | CI-95% |
|---|---|---|---|
| C. parapsilosis sensu stricto | 24 | 0.96 | 95.7-96.3% |
| C. orthopsilosis | 0 | 0 | 0 |
| C. metapsilosis | 0 | 0 | 0 |
| Others | 1 | 0.04 | 4-12% |
| Total | 25 | 1 |
Table 4: Distribution of species of the C. parapsilosis complex in the sample analyzed.
Of the strains characterized as C. parapsilosis sensu stricto, 79.2% (19/25) were from immunocompromised patients (oncohematology, diabetes and pre-term newborns). The difference was statistically significant, with prevalence ratio (PR) 0.26, which means that immunocompetent subjects are less likely to develop invasive candidiasis by Candida parapsilosis sensu stricto than are immunocompromised patients, who would be more vulnerable to it (Table 5).
Figure 1: Electrophoretic run of PCR products from 17 samples from different sites of the oral cavity. A binder pattern of 379 bp is observed.
Note: WM: Weight marker; C+: Positive control; C-: Negative control; samples 1,2,3,4,5,6,7,9,10,11,12,13,14,15,16,17 compatible with species C. parapsilosis sensu stricto. Sample 8 negative for species C. parapsilosis sensu stricto with primers CPAR-CPAF
| Immune Status | C. parapsilosis sensu stricto N (%) |
|---|---|
| Immunocompetent | 5 (20.8) |
| Immunocompromised | 19 (79.2) |
| Total | 24 (100) |
p-Fisher: 0.000059653 PR: 0.26
Table 5: Distribution of C. parapsilosis sensu stricto strains according to host immune status.
The species-specific clinical breakpoints proposed by the CLSI Subcommittee in 2012 showed that of the 24 strains derived from blood and reconfirmed as C. parapsilosis sensu stricto, 20 (83.3%) were sensitive to FZ, while 100% were sensitive to VZ. For echinocandins, 23 out of 24 (95.8%) were sensitive to CAS, while 100% were sensitive to MICA (Table 6).
| Drug | S N (%) |
SI N (%) |
SDD N (%) |
R N (%) |
WT N (%) |
non WT N (%) |
|---|---|---|---|---|---|---|
| FZ | 20 (83.3) | - | 3 (12.5) | 1 (4.2) | 20 (83.3) | 4 (16.7) |
| VZ | 24 (100) | - | - | - | 24 (100) | 0 |
| CAS | 23 (95.8) | - | - | 1 (4.2) | 21 (87.5) | 3 (12.5) |
| MICA | 24 (100) | - | - | - | 24 (100) | 0 |
| AB | ND | ND | ND | ND | 24 (100) | 0 |
| FC | ND | ND | ND | ND | 0 | 24 (100) |
Note: S=Susceptible; SI=Intermediate; SDD=Susceptible Dose-dependent; R=Resistant; WT=Wild Type; non WT=Non-Wild Type; ND=Not Determined.
Table 6: Distribution of response phenotypes to each drug tested on the study strains.
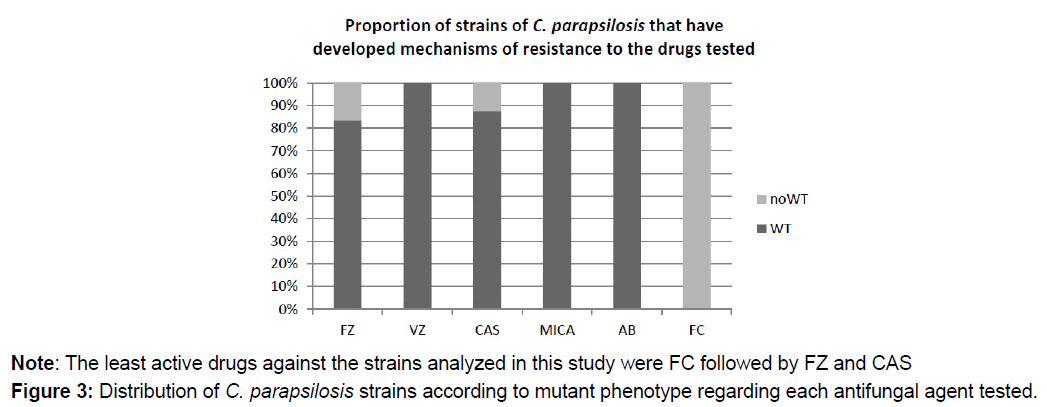
No results can be provided for AB and FC, because there is no clinically validated breakpoint for these drugs in C. parapsilosis. However, according to epidemiological cutoff value (ECV), 100% of the strains were wild type to AB; but non-wild type phenotype to FC, i.e., they have developed resistance mechanisms against FC (Figure 3).
| Drug | CIM50 (mg/ml) | CIM90 (mg/ml) | Range CIM (mg/ml) | Frequency of Resistance |
|---|---|---|---|---|
| FZ | = 1 | 4 | = 1-64 | 16.70% |
| VZ | = 0.12 | = 0.12 | = 0.12 | 0% |
| CAS | 1 | 1 | 1 | 12.50% |
| MICA | 0.5 | 0.5 | 0.5 | 0% |
| AB | = 0.25 | 0.5 | 0.25-0.5 | ND |
| FZ | 1 | 1 | 1 | ND |
Note: ND: Not Determined.
Table 7: CIM50 and CIM90 values for each drug tested on the 24 strains evaluated.
5. Discussion
The results of this study suggest that C. parapsilosis sensu stricto is more likely to be retrieved than are other species in the complex from blood cultures from Argentinean subjects. This is consistent with contributions from other authors (Podesta [13]; Rodríguez [14] and Madeot [15]), who analyzed the epidemiology of this complex in samples from patients with invasive disease Argentina, reporting recovery frequency at nearly 90% for C. parapsilosis sensu stricto; with C. metapsilosis being the second most frequently isolated species in the complex from blood samples. However, the number of isolates used in the papers cited, as in ours, is limited, so the same analysis should be performed on a larger sample in order to determine the behavior of the rarer species in the complex in invasive samples from the Argentinean population.
Comparison of C. parapsilosis sensu stricto strains to patient immune status at the time of sampling shows that the probability of recovering C. parapsilosis sensu stricto from blood is greater in immunocompromised patients. This is in contrast to Constante et al [16], who found other species from the complex in patients with immune deterioration. Nevertheless, this should be analyzed in a larger number of isolates using a prospective design.
By literature it is known that C. parapsilosis sensu stricto isolates are less susceptible than C. orthopsilosis and C. methapsilosis isolates to some antifungal agents used for treating candidiasis, such as Amphotericin (AB), Fluconazol (FZ), Itraconazole (IZ) and Caspofungin (CAS). Considering the azole derivatives analyzed in our study, FZ was the least active, with 4 C. parapsilosis sensu stricto strains having little susceptibility to it. In agreement, Silva et al. [4] found MIC=16 mg/mL for FZ for a C. parapsilosis sensu stricto isolate, and Gomez-Lopez et al. [17] reported similar results. In contrast, Ataides et al. [6] and Ruiz et al. [18] reported resistance of a C. parapsilosis sensu stricto isolate to IZ but not to FZ. In this regard, Van Asbeck et al.[19] suggest that the differences in susceptibility to FZ may also reflect the different affinities of azoles to the key enzyme that synthesizes ergosterol, 14-α-demethylase, or to other enzymes in this pathway. Interestingly, MIC50 and MIC90 values for both azoles (Table 7) found for the 24 study strains are comparatively higher than those reported for other regions such as Turkey and Spain, reflecting geographic variability in the response of these species in this complex to antifungal drugs [2,20].
For echinocandins, only one isolate was resistant to CAS in this study. This is in agreement with the worldwide trend whereby several studies report that caspofungin MIC values for C. parapsilosis sensu stricto are higher than those for the other two species in the complex. In addition, MIC50 and MIC90 values for both echinocandins (Table 7) were lower than those reported in other parts of the world such as Turkey and Spain, but similar to those reported by Cantón et al. [21].All the isolates were susceptible to micafungin.
Based on ECV, all isolates had non-mutant phenotype for amphotericin B. This result is similar to those reported in other studies [2,22]. However, Ataides et al. [6] and Lockhartet al. [5] reported resistance of C. parapsilosis sensu stricto to AB. The response of species in the Candida parapsilosis complex to AB varies considerably among regions, which may be determined by intra-species genotype variability.
With regard to response to flucytosine, in our study, 100% of the isolates showed MIC=1mg/mL of the drug. Like AB, there is no consensus on the breakpoint of flucytosine for Candida parapsilosis sensu stricto. However, considering the ECV categorization suggested by Pfaller and Diekema in 2012, we found that 100% of the strains were resistant to FC, since according to the ECV, MIC £ 0.5 mg/ml for FC corresponds to a WT strain, while MIC>0.5ug/ml indicates a mutant strain which has developed mechanisms of resistance [12]. Nevertheless, the British Society for Mycopathology establishes FC-“sensitive” isolates as those with MIC £ 1 mg/ml [23]. The MIC value for FC found in our study is high compared to those reported by Silva et al. [4], Miranda et al. [2] and Cantón et al. [21] for studies conducted in Europe. In India, Bhattet al. [24] found a high rate of resistance to FC in C. parapsilosis isolates. C. orthopsilosis or C. metapsilosis were not recovered; therefore it was not possible to establish differences in the response profiles among the 3 species in this complex.
6. Conclusion
C. parapsilosis sensu stricto is the most frequently identified species in the complex in blood cultures from Argentinean patients with candidemia, being more likely to be found in immunocompromised patients.
There is geographic variability for the response of C. parapsilosis sensu stricto strains to the antifungal agents most commonly used in clinical practice.
Flucytosine is not very active against pathogenic strains of C. parapsilosis sensu stricto. However we suggest analyzing the action of this drug with two methods of sensitivity in vitro, and in a larger sample size.
References
- Ge Y, Boekhout T, Zhan P, et al. (2012). Characterization of the Candida parapsilosis complex in East China: Species distribution differs among cities. Med Mycol. 50: 56-66.
- Miranda I, Eraso E, Hernández J, et al. (2011). Prevalence and antifungal susceptibility patterns of new cryptic species inside the species complexes Candida parapsilosis and Candida glabrata among blood isolates from a Spanish tertiary hospital. J Antimicrob Chemother. 66: 2315–2322.
- Tavanti A, Davidson A, Gow N, et al. (2005). Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J Clin Microbiol. 43: 284-292.
- Silva A, Miranda I, Lisboa C, et al. (2009). Prevalence, distribution and antifungal susceptibility profiles of Candida parapsilosis, C. orthopsilosis and C. metapsilosis in a Tertiary Care Hospital. J Clin Microbiol. 47: 2392–2397.
- Lockhart S, Messer S, Pfaller M, et al. (2008). Geographic distribution and antifungal susceptibility of the newly described species Candida orthopsilosis and Candida metapsilosis in comparison to the closely related species Candida parapsilosis. J Clin Microbiol. 46: 2659-2664.
- Ataides F, Costa C, Souza HL, et al. (2015). Molecular identification and antifungal susceptibility profiles of Candida parapsilosis complex species isolated from culture collection of clinical samples. Rev Soc Bras Med Trop. 48: 454-459.
- Ochiuzzi M, Arechavala A, Guelfand L, et al. (2014). Evaluación de las tarjetas AST-Y01 del sistema Vitek 2 para determinar la sensibilidad a antifúngicos de levaduras del género Candida. Revista Argentina de Microbiología. 46: 111-118.
- Borghi E, Iatta R, Sciota R, et al. (2010). Comparative evalutation of the Vitek 2 yeast susceptibility test and CLSI broth microdilution reference method for testing antifungal susceptibility of invasive fungal isolates in Italy: The GISIA3 study. J Clin Microbiol. 48: 3153-3157.
- Mujica M, Finquelievich J, Jewtuchowicz V, et al. (2004). Prevalence of Candida albicans and Candida non-albicans in clinical samples during 1999-2001. Rev Argent Microbiol. 36: 107-112.
- Silva S, Negri M, Henriques M, et al. (2012). Candida glabrata, Candida parapsilosis and Candida tropicalis: Biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol Rev. 36.
- Asadzadeh M, Ahmad S, Al-Sweih N, et al. (2009). Rapid molecular differentiation and genotypic heterogeneity among Candida parapsilosis and Candida orthopsilosis strains isolated from clinical specimens in Kuwait. J Med Microbiol. 58: 745-752.
- Pfaller M, Diekema D. (2010). Progress in antifungal susceptibility testing of Candida spp. by use of clinical and laboratory standards institute broth microdilution methods, 2010 to 2012. J Clin Microbiol. 50: 2846–2856.
- Podesta M, Funes P, Amigot S, et al. (2016). Molecular identification and antifungal susceptibility profiles of Candida parapsilosis complex species isolated from clinical samples in a Argentina Hospital. Libro de resúmenes del Forum on fungal infection in the clinical practice, Chile.
- Rodriguez L, Mayo S, Sellart G, et al. (2015). Distribución, sensibilidad a antifúngicos y biopelículas del complejo parapsilosis en candidemia hospitalaria. Libro de resúmenes del Forum on fungal infection in the clinical practice. Argentina.
- Medeot M, Sartori L, Abiega C, et al. (2015). Estudio de Candidemia en Córdoba Argentina. Libro de resúmenes del Forum on fungal infection in the clinical practice. Argentina.
- Constante C, Monteiro A, Alves A, et al. (2014). Different risk factors for candidemia occur for Candida species belonging to the C. parapsilosis complex. Med Mycol. 52: 403-406.
- Lopez GA, Izquierdo AA, Rodriguez D, et al. (2008). Prevalence and susceptibility profile of Candida metapsilosis and Candida orthopsilosis: Results from population-based surveillance of candidemia in Spain. Antimicrob Agents Chemother. 52: 1506-1509.
- Ruiz L, Khouri S, Hahn R, et al. (2013). Candidemia by species of the Candida parapsilosis complex in children's hospital: Prevalence, biofilm production and antifungal susceptibility. Mycopathol. 175: 231-239.
- Van Asbeck E, Clemons K, Martínez M, et al. (2008). Significant differences in drug susceptibility among species in the Candida parapsilosis group. Diagn Microbiol Infect Dis. 62: 106-109.
- Tosun I, Akyuz Z, Guler N, et al. (2012). Distribution, virulence attributes and antifungal susceptibility patterns of Candida parapsilosis complex strains isolated from clinical samples. Med Mycol.
- Canton E, Pemán J, Quindós G, et al. (2011). Prospective multicenter study of the epidemiology, molecular identification, and antifungal susceptibility of Candida parapsilosis, Candida orthopsilosis and Candida metapsilosis isolated from patients with Candidemia. Antimicrob Agents Chemother. 55: 5590-5596.
- Córdoba S, Vivot W, Bosco-Borgeat M, et al. (2011). Species distribution and susceptibility profile of yeasts isolated from blood cultures: Results of a multicenter active laboratory-based surveillance study in Argentina. Revista Argentina de Microbiología. 43: 176-185.
- British Society for Mycopathology. (1984). Laboratory methods for flucytosine (5-fluorocytosine). Report of a working group of the British Society for Mycopathology. J Antimicrob Chemother. 14: 1-8.
- Bhatt M, Sarangi G, Paty B, et al. (2015). Biofilm as a virulence marker in Candida species in nosocomial blood stream infection and its correlation with antifungal resistance. Indian J Med Microbiol. 33: 112-114.
- White TJ, Bruns TD, Lee SB, et al. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds). PCR Protocols: A Guide to Methods and Applications. San Diego, CA: Academic Press Inc., pp: 315-322.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences