Bioreduction of uranium using native Enterococcus faecalis and Lactococcus garvieae
Faezeh Fatemi*, Samaneh Jahani, Nooshin Hashemi
Faezeh Fatemi1*,Samaneh Jahani2 and Nooshin Hashemi3
1Materials and Nuclear Fuel Research Center, Research Center for Nuclear Science and Technology, Iran, Tehran
2Department of Microbiology, Faculty of Science, Qom Branch, Islamic Azad University, Qom, Iran
3Department of Biotechnology, Faculty of Life Science, Alzahra University, Tehran, Iran
Received date January 21, 2019; Accepted date February 13, 2019; Published date February 28, 2019
Citation: Fatemi F, Jahani S, Hasheemi N. Bioreduction of uranium using native Enterococcus faecalis and Lactococcus garvieae Electronic J Biol, 15:1
Abstract
The aim of this study is the isolation of the native bacteria from uranium mine with the potency of uranium removal in waste via reduction. The isolates is described as E. faecalis FJ4, L. garvieae FJ5, L. garvieae FJ6 and E. faecalis FJ7 seeing that Grampositive, cocci and facultative anaerobic bacteria. The isolates in the presence of U (VI) aqueous solutions led to formation of a black precipitate under anaerobic condition. ICP analysis of the solution in 2nd and 4th days of incubation indicated successful uranium removal of the solution. Moreover, analysis of the precipitate using UV-vis confirmed the reduction of U (VI) to U (IV). As a conclusion, the native isolates showed the ability of uranium removal from the contaminated wastes with uranium.
Keywords
Isolation; Characterization; Enterococcus faecalis; Lactococcus garvieae; Uranium reduction.
1. Introduction
The diffusion of radionuclides from radionuclide contaminated sites and their mobility in the environment is a subject of public concern [1]. Radioactive waste microbiology started with the understanding that bacteria could be present in disposal of waste and the effects of microbial activity could have deep influences on waste containment [2]. Bacteria can potentially affect radionuclide migration using various processes. Bacteria decline radionuclide migration by reduction, sorption, altering bulk pore water chemistry, producing organic complexing ligands and by direct accumulation onto or into cells [3]. Furthermore, bacteria can also cause corrosion and hence potentially affect the longevity of the metal waste containers in a repository [4].
Uranium as natural sources of radioactivity is present in earth's crust at concentration of 1.8 ppm [1]. As explained above, bacterial reduction is an effective way for decline radionuclide migration (including uranium). This process is a microbial reaction between dissolved uranium, U (VI), and some kinds of bacteria in which electrons are transported from an electron donor to U (VI), as final electron accepter. Moreover, bioreduction is mediated by catalytic intermediary of reducing enzymes of bacteria that results in uranium reduction to the non-soluble form, U (IV), [5, 6].
The reduction of soluble U (VI) to insoluble U (IV) has been suggested as a device for inhibiting the migration of this toxic metal with ground water [5, 7]. Bacterial reduction of U (VI) is a suitable method that was first reported in crude extracts from Micrococcus lactilyticus by assaying the consumption of hydrogen which was dependent on the existence of U (VI) [8]. Nowadays more than 25 species of phylogenetically diverse prokaryotes are identified that be involved in the reduction of radionuclides [9]. A number of these prokaryotes conserve energy from U (VI) reduction for their growth, and some of them reduce uranium without energy gain [5, 10, 11, 12, 13]. Studies about bioreduction by TEM analysis indicated that the precipitated uraninite has been located in the periplasm and outside of Gram-negative and Grampositive bacterial cells [14, 15, 16]. The research suggested that U (VI) complexes do not generally have access to intracellular enzymes [6]. Microorganisms are able to precipitate metals and radionuclides as carbonates and hydroxides by localized alkalinization at the cell surface [17]. On the other hand, metals can precipitate with enzymatically-generated ligands, e.g., phosphate, sulfide, oxalate, etc. [18, 19, 20].
The aim of this study is the isolation and characterization of native bacteria with novel ability of uranium reduction from Bandar Abbas uranium mine, Iran. For this purpose, after isolation process, the ability of isolates for uranium removal from aqueous solutions was investigated and the mechanism of removal was studied using UV spectrums.
2. Materials and Methods
2.1 Bacteria isolation
Sampling: Four samples collected from the area depths of water, sledge and soil of Bandar Abbas uranium mine, Iran. The samples were stored anaerobically at 4°C before it was shipped to the laboratory.
Strain purifications: Anaerobic techniques [21] were used for the enrichment and isolation of the strains. The vials were purged with oxygen free gas for making anaerobic atmospheres. The vials were containing 9 ml of egg yolk medium and 0.5 g of the samples. Modified egg yolk medium contained proteose peptone (40 g/liter), sodium chloride (2.0 g/liter), Na2HPO4 (5.0 g/liter), KH2PO4 (5.0 g/liter), MgSO4 (0.1 g/liter), glucose (2.0 g/liter), egg yolk suspension (100 mL) and demineralized water (1000 mL). All cultures were incubated at 30°C. After 10 days, the cultures were serially diluted with the same anaerobic medium. The culture in the last positive dilution was purified by repeated plating onto the anaerobic agar plates with egg yolk agar medium.
2.2 Bioreduction experiment
Isolate cultivations: The isolates cultivated in Tryptic Soy Broth (TSB) medium under anaerobic conditions. The pH mediums was adjusted to 7.26 and incubated to 30°C for 24 hours. The cell samples were harvested by centrifuge at 4500 rpm for 25 minutes and then washed with sodium bicarbonate solution (2 g L–1). The washed cells were suspended in the sodium bicarbonate solution as inoculums and the pH was adjusted to 7.26.
Bioreduction solutions: The medium used for bioreduction was contained the following solutions: salts including (g L–1) NH4Cl (0.25), KCl (0.5), CaCl2.2H2O (0.15), NaCl (1.0), MgCl2.6H2O (0.62) and vitamins including (mg L–1) p-aminobenzoic acid (0.05), thiamine-HCl (0.02), pyridoxine-HCl (0.1), cyanocobalamin (0.001) and trace minerals including (mg L–1) MnCl2.4H2O (0.1), CoCl2.6H2O (0.12), ZnCl2 (0.07), H3BO3 (0.06), NiCl2.6H2O (0.025), CuCl2.2H2O (0.015), Na2MoO4.2H2O (0.025), FeCl2.4H2O (1.5). In following, 10 mM of sodium lactate (50% syrup) was added as electron donor. The uranium (up to 0.5 mM) was added as the species uranyl acetate. The pH was adjusted by sodium bicarbonate [22].
2.3 Anaerobic cultivations
After pH adjustment, 9 mL volumes of mediums were transferred to 25 mL Balch-type vials. Then, the vials were urged with a mixed gas (90% N2 and 10% CO2) by a stainless steel cannula as nozzle for 5 minutes and immediately the sealing of vials carried out with rubber and aluminum caps. In following, hydrogen gas from a continuous H2-generator was injected into the vials by a thin needle up to pressure +0.6 bars. Presence of H2 gas in the vials at a positive pressure guarantees that any remaining or probable oxygen leakage toward inside, will be vanished via water forming reaction [7,23]. After that, one mL of the inoculums was injected to each anaerobic vials containing 9mL reduction solution which were incubated at 30°C. It should be noted that, tow control samples including cell-free control for abiotic precipitation and U-free medium control for assuring uranium precipitation - not the cells- were provided.
2.4 Analysis of U (IV) and U (VI)
Soluble uranium (IV) was determined by Inductively Coupled Plasma Spectrometry (ICP, Perkin Elmer Optima 2000 DV). The culture samples were transferred to a centrifuge tube under anaerobic conditions (glove box). Then, the pH was adjusted to 11 with 1 N NaOH to precipitate the uranium and the cultures were centrifuged at 4500 rpm for 20 min. The supernatant discarded, and the pellet resuspended in 2 mL of a 10 mM citric acid solution to extract the uranium species. The solutions were filtered by 0.45 μm membrane filters and analyzed by UV-visible light (UV-vis) spectrophotometry to determine the absorption spectra of uranium (IV) [24].
2.5 Characterization of uranium reducer strains
Phenotypic characteristics: The Gram staining and microscopic observations of the isolates were carried out in exponential growth phase. Diagnosis obligate or facultative anaerobic of isolates was carried out using bacterial culture under aerobic and anaerobic conditions.
Genotypic characteristics: The DNAs were extracted from the isolates using DNA extraction kit (Cinna Clone) according to the manufacturer's instructions. The primers used for PCR amplification of the 16S rRNA sequences were 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). The PCR amplification was carried out as follows: 50 ng of genomic DNA, 0.75 μL MgCl2, 0.5 μL dNTPs, 1 μL each of sense and antisense primers, 2.5 μL of 10X reaction buffer (Thermo Scientific) and 0.4 μL of pfu DNA polymerase (Vivantis) in a total volume of 25 μL. The reaction mixtures were amplified in a thermal cycler (PeQ lab, England) by use of the following program: 95°C for 3 min, followed by 30 cycles consisting of denaturation at 95°C for 30 s, annealing at 50°C for 30 s and extension at 72°C for 90 s. The final extension step was 72°C for 7 min. After that, the PCR products were purified and sequenced. Finally, sequence alignment was performed by BioEdit software and were searched for homology by BlastN at the NCBI server (https:// www.ncbi.nlm.nih. gov/BLAST/Blast) and submitted to Genbank. Also, a phylogenetic tree was constructed with MEGA 5 software based on the 16S rRNA sequences of other closer strains.
Results and Discussion
3.1 Isolation and purification
The bacterial cells have adapted to the environments contaminated with toxic metals, in order to grow and survive and use the essential metals to drive their metabolic machinery, and also protect themselves against toxic elements such as radionuclides through different mechanisms such as bioreduction, biosorption and etc. [25, 26, 27]. So, improved remediation will be achieved when the isolation of the bacteria was performed from the native environment.
In this study, the samples obtained from water, sledge and soil of Bandar Abbas uranium mine, Iran, were inoculated onto modified egg yolk medium. After 10 days of incubation at 30°C, the bacteria were developed in the purging vials which were oxygen free gas. The grown cells of bacteria were inoculated onto solid medium under anaerobic conditions. Such ordinary purification procedures were repeated several times, finally pure cultures were obtained. The results indicated that, the colonies of all samples were smooth, spherical and milky in color. After isolation, in order to coding the strains, they were called FJ4, FJ5, FJ6 and FJ7.
3.2 Ash content
The ash content (%) of the honey samples fall within the range presented in Table 1 (0.020 to 1.028% for commercial honey). The minimum value for ash contents was 0.061% and the maximum was 0.434%. Sample 8 had the highest inorganic (ash) content but were within the legal parameters, as was the corn glucose sample. Correctly processed pure honey presents low ash contents, with a maximum of 1%. This parameter is therefore commonly used to identify irregularities such as the presence of insects, paint residue, pieces of wood or wax, indicative of lack of proper filtration or hygiene. Brazilian legislation permits a maximum of 0.6 g/100 g (or 0.6%) for floral honey. For honeydew and its mixtures with honey, up to 1.2 g/100 g is accepted [16].
| Name of samples | U(VI) concentration after 2 days incubation | U(VI) concentration after 4 days incubation |
|---|---|---|
| FJ4 | 24 ppm | >1 ppm |
| FJ5 | 27 ppm | >1 ppm |
| FJ6 | 27 ppm | >1 ppm |
| FJ7 | 46 ppm | 15 ppm |
| Cell-free Control | 127 ppm | 68 ppm |
| U- free medium | 0 ppm | 0 ppm |
Table 1. The results of solution analysis by ICP method.
3.3 Uranium bioreduction
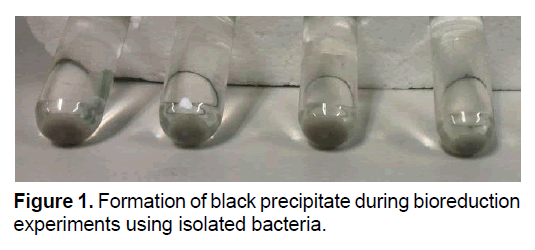
To identify the ability of the isolates in the uranium reduction, inoculums were injected into the anaerobic vials containing reduction solution and incubated at 30°C. The results showed that, a dark precipitate formed during incubation time, while the yellow color of solution disappeared due to transportation of uranium from the solution phase to the precipitate. These happened in all vials except control samples. Figure 1 showed the morphological feature of the precipitates of uranium bioreduction.
Analysis of the solution in 2nd and 4th days of incubation using ICP, confirmed the uranium removal of the solution showed in Table 1 pointing to that the isolates are capable of uranium removal. The results indicated that, the concentration of U(VI) in solutions was 24, 27, 27, 46, 127 and 0 ppm for FJ4, FJ5, FJ6, FJ7 strains, cell-free control and U-free medium control in 2nd day, while the amount was reduced to >1, >1, >1, 15, 68 and 0 ppm in 4th day, respectively.
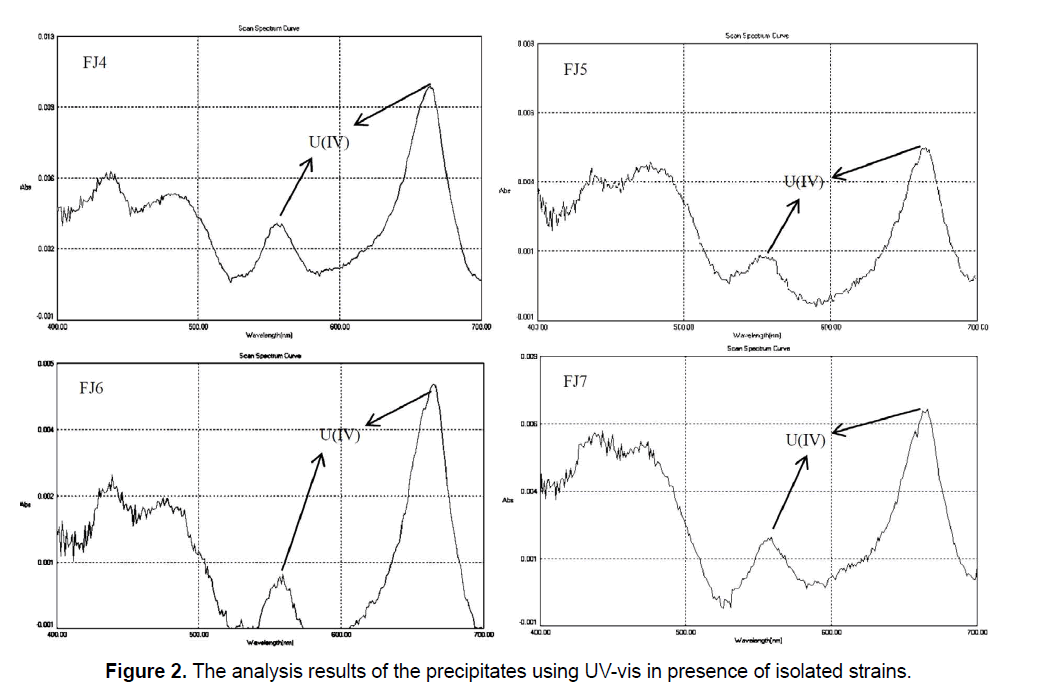
In addition, the absorption spectra determination of uranium (IV) in precipitates showed the major and minor peaks at about 662 nm and 560 nm, respectively (Figure 2). Gao and Francis (Gao and Francis 2008) [24] reported that respective UV-vis spectra for U (IV) is 662 nm and 560 nm that confirm our results in the present study. According to Figure 2, it was clear that U (VI) was removed from the solution and converted to U (IV) in the precipitate due to presence and activity of the bacterial cells. The significant decrease in solubility beside the reduction of U (VI) to U (IV), producing the insoluble uraninite, has been viewed as a potential mechanism for clearing of environmental uranium contamination.
In the past years, it has been tightly established that some species of bacteria show this ability.
It can be deduced that removal of U during experiments can be attributed primarily to reductive precipitation of UO2, and not to adsorption onto the bacterial cell membranes. Lovley, 1993 [28] explained that bacterial dissimilatory U (VI) reduction, that leading to U (IV) precipitation as UO2, has been proposed as an alternative means of promoting U mineralization. Also, initial uranium bioreduction experiments used X-ray diffraction (XRD) and transmission electron microscopy (TEM) to identify the black mineral precipitate formed as uraninite UO2 [14, 29].
The following equation is summary of uranium reduction process by bacteria [7].

It should be noted that, uranium bio-reduction is an anaerobic process. Thus, in the absence of oxygen, U (VI) plays the role of electron acceptor instead of oxygen [6]. Uranium bio-reduction is a microbial reaction between U (VI) and some kinds of bacteria in which electrons are transported from an electron donor to U (VI), as final electron accepter. This process is mediated by catalytic intermediary of reducing enzymes of the bacteria that results in uranium reduction to the non-soluble form, U (IV). The researchers reported that reduction of soluble U(VI) to insoluble U(IV) has been proposed as a mechanism for preventing the migration of this toxic element with ground water [7, 8].
There are different mechanisms for different species in order that cells transfer electrons from the electron donor to an electron acceptor such as U (VI) that is debated. Although, U(VI) can be significantly soluble in certain environmental conditions and therefore may diffuse into the direct contact with a cell. Bacteria are also capable of transferring electrons to solid electron acceptors such as sorbed/precipitated U (VI). Therefore, bacteria must have evolved mechanisms for transporting electrons from the central metabolism to the outside of the cytoplasmic membrane, the periplasm and the outer membrane and potentially extracellular space. These include the use of electron-carriers such as cytochromes or flavins or through the expression of conductive cell surface appendages such as pili. It should be noted that, the mechanisms of U (VI) bioreduction are not yet fully resolved, especially the significance of the role played by pili and electron shuttles [30].
3.4 Characterization of the isolates
Phenotypic characterization: The compound microscopic observations of the isolated strains revealed that, all strains were Gram-positive and facultative anaerobic. Furthermore, two of the samples were cocci in clusters, short chains, diplococcic, single cocci and another isolates were ovoid coccus, occurring in pairs and short chains.
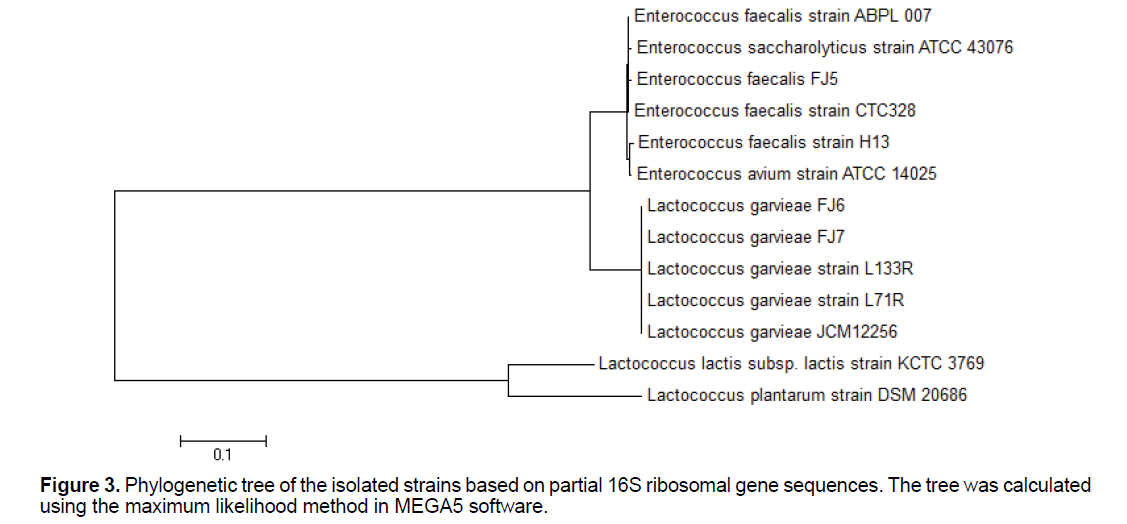
Genotypic characterization: Maximum likelihood tree building is shown in Figure 3. The data presented showed that L. garvieae and E. faecalis, which both have ability to uranium reduction, can be easily differentiated by the proposed PCR assay. The PCR assay that we have developed is a convenient, fast, and simple technique that can be accomplished in 5 h and that can be applied to the specific identification of L. garvieae and E. faecalis.
The PCR assay resulted in the amplification of a band of 1465 bp in size detected for all the strains. Following sequences, BLAST search, alignments and analysis of the 16S rRNA gene sequences indicated that the 2 bacterial strains feel righted to phylogenetically different groups. Strain FJ4 had 16S rRNA gene similarities of 99%-98% belonged to the members of the genus Enterococcus, with the highest similarity to E. faecalis. Strain FJ5 had 16S rRNA gene similarities of 99%–97% belonged to the members of the genus Lactococcus, with the highest similarity to L. garvieae. Strain FJ6 had 16S rRNA gene similarities of 99%–98% related to the members of the genus Lactococcus, with the highest similarity to L. garvieae. Strain FJ7 had 16S rRNA gene similarities of 98%-97% belonged to the members of the genus Enterococcus, with the highest similarity to E. faecalis. Nucleotide sequences of examined strains gene fragments were recorded in NCBI database (accession numbers of deposits KX671994.1, KX671995.1, KX671996.1 and KX671997.1 related to Enterococcus faecalis strain FJ4, Lactococcus garvieae strain FJ5, Lactococcus garvieae strain FJ6 and Enterococcus faecalis strain FJ7, respectively.
Many researchers have been isolated L. garvieae and E. faecalis from various environments [31, 32, 33], but there is no report about the isolation of these bacteria from uranium mine. In addition, according to our researches, no evidence exists regarding the ability of L. garvieae for uranium reduction. It should be noted that, there is just a thesis about existence of E. faecalis capable of uranium reduction [34]. In the present study, we succeeded to isolate native bacteria with the ability of uranium reduction, from uranium mine, under anaerobic condition for the first time.
Conclusion
The results of this study showed the successful isolation of native uranium reducer bacteria from Iranian uranium mine. In the present study, we isolated 4 native strains capable of uranium reduction under anaerobic condition. Analysis of the precipitate by UV-vis confirmed that the mechanism of uranium removal is microbial reduction from U(VI) to U(IV). In addition, the results of the characterization revealed that all strains were Gram-positive, facultative anaerobic and cocci. Sequencing of 16S rRNA indicated that, the isolates are members of the genus Enterococcus and Lactococcus. Finally, we reported for the first time that the native strains including E. faecalis FJ4, L. garvieae FJ5, L. garvieae FJ6 and E. faecalis FJ7 were found as able native bacteria for microbial reduction of U(VI) from aqueous solution.
Acknowledgments
We thank Dr. Hassan Zare Tavakoli a lot for help in sampling
References
- Merroun ML, Selenska-Pobell S (2007). Transmission Electron Microscope Analysis of Eu(III) accumulated by Bacillus sphaericus JG-A12, Report FZD. 459: 44.
- West J, Mckinley IG, Chapman NA. (1982). Microbes in deep geological system and their possible influence on radioactive waste disposal. Radioact Waste Manage Nucl Fuel Cycle. 3: 1-15.
- West JM, Mckinley IG, Stroes-Gascoyne S. (2002). Microbial effects on waste repository materials. In: Keith-Roach, M., Livens, F. (Eds.), Interactions of microorganisms with radionuclides. Elsevier Sciences, Oxford, UK. 255-277.
- Stroes-Gascoyne S, Schippers A, Schwyn B, et al. (2007). Microbial community analysis of opalinus clay drill core samples from the Mont Terri Underground Research Laboratory, Switzerland. Geomicrobiol. 24: 1-17.
- Lovley DR, Phillips EJP, Gorby YA, et al. (1991). Microbial reduction of uranium. Nature. 350: 413-416.
- Wall JD, Krumholz LR. (2006). Uranium reduction. Annu. Rev Microbiol. 60: 149-166.
- Gorby YA, Lovley DR. (1992). Enzymatic uranium precipitation. Environ Sci Technol. 26: 205-207.
- Woolfolk CA, Whiteley HR. (1962). Reduction of inorganic compounds with molecular hydrogen by Micrococcus lactilyticus. I. Stoichiometry with compounds of arsenic, selenium, tellurium, transitions and other elements. J Bacteriol. 84: 647-658.
- Suzuki Y, Suko T. (2006). Geomicrobiological factors that control uranium mobility in the environment: Update on recent advances in the bioremediation of uranium-contaminated sites. J Mineral Petrol Sci. 101: 299-307.
- Francis AJ, Dodge CJ, Lu F, et al. (1994). XPS and XANES studies of uranium reduction by Clostridium sp. Environ Sci Technol. 28: 636-639.
- Kashefi K, Lovely DR. (2000). Reduction of Fe(III), Mn(IV), and toxic metals at 100 C by Pyrobaculum islandicum. Appl Environ Microbiol. 66: 1050-1056.
- Lloyd JR, Macaskie LE. (2002). Biochemical basis of microbe-radionuclide interactions. In: Keith-Roach, M., Livens, F. (Eds.), Interactions of Microorganisms with Radionuclides. Elsevier Sciences, Oxford, UK. 313-342.
- Pietzsch K, Hard BC, Babel W. (1999). A Desulfovibrio sp. capable of growing by reducing U(VI). J Basic Microbiol. 39: 365-372.
- Lovley DR, Phillips EJP. (1992). Reduction of uranium by Desulfovibrio desulfuricans. Appl Environ Microbiol. 58: 850-856.
- Xu H, Barton LL, Zhang P, et al. (2000). TEM investigation of U (VI) and Re(VII) reduction by Desulfovibrio desulfuricans, a sulfate reducing bacterium. Mater Res Soc Symp Proc. 608: 299-304.
- Liu H, Fang HHP. (2002). Characterization of electrostatic binding sites of extracellular polymers by linear programming analysis of titration data. Biotechnol Bioeng. 80: 806-811.
- Van Roy S, Peys K, Dresselaers T, et al. (1997). The use of an Alcaligenes eutrophus biofilm in a membrane bioreactor for heavy metals recovery. Res Microbiol. 148: 526-528.
- Yong P, Macaskie LE. (1995). Role of citrate as complexing ligand which permits enzymatically-mediated uranyl ion bioaccumulation. Bull. Environ Contam Toxicol. 54: 892-899.
- Macaskie LE, Empson RM, Cheetham AK, et al. (1992). Uranium bioaccumulation by a Citrobacter sp. as a result of enzymically mediated growth of polycrystalline HUO2PO4. Science. 257: 782-784.
- Joeng BC, Hawes C, Bonthrone KM, et al. (1997). Localization of enzymatically enhanced heavy metal accumulation Citrobacter sp. and metal accumulation in vitro by liposomes containing entrapped enzyme. Microbiology. 43: 2497-2507.
- Tavakoli H, Sepehrian H, Semnani F, et al. (2013). Recovery of uranium from UCF liquid waste by anion exchange resin CG-400: breakthrough curves, elution behavior and modeling studies. Ann Nucl Energy. 54: 149-153.
- Czerwinski KR, Polz MF. (2008). Uranium enrichment using microorganisms, Massachusetts Institute of Technology, Boston, MA.
- Mullen LM. (2007). Bacterial influence on uranium oxidation reduction reactions: implications for environmental remediation and isotopic composition, PhD Thesis, Massachusetts Institute of Technology, Cambridge, MA.
- Gao W, Francis A J. (2008). Reduction of uranium (VI) to uranium (IV) by Clostridia. Appl Environ Microb. 74: 4580-4584.
- Merroun ML, Raff J, Rossberg A, et al. (2005). Complexation of uranium by cells and S-layer sheets of Bacillus sphaericus JG-A12. Appl Environ Microbiol. 71: 5542-5553.
- Nedelkova M, Merroun ML, Rossberg A, et al. (2007). Microbacterium isolates from the vicinity of a radioactive waste depository and their interactions with uranium. FEMS Microbiol Ecol.59: 694-705.
- Suzuki Y, Banfield J. (2004). Resistance to, and accumulation of, uranium by bacteria from a uranium-contaminated site. Geomicrobiol J. 21: 113-121.
- Lovley DR. (1993). Dissimilatory metal reduction Annu. Rev. Microbiol. 47: 263.
- Abdelouas A, Lu Y, Lutze W, et al. (1998). Reduction of U (VI) to U (IV) by indigenous bacteria in contaminated ground water. J Contam Hydrol.35: 217-233.
- Pfeffer C, Larsen S, Song J, et al. (2012). Filamentous bacteria transport electrons over centimetre distances. Nature. 491: 218-221.
- Evans JJ, Klesius PH, Shoemaker CA. (2009). First isolation and characterization of Lactococcus garvieae from Brazilian Nile tilapia, Oreochromis niloticus (L.), and pintado, Pseudoplathystoma corruscans (Spix & Agassiz). J Fish Dis. 32: 943-951.
- Eldar A, Goria M, Ghittino C, et al. (1999). Biodiversity of Lactococcus garvieae strains isolated from fish in Europe, Asia, and Australia. J Appl Environ Microbiol. 65: 1005-1008.
- Mahmoudpour A, Rahimi S, Sina M, et al. (2007). Isolation and identification of Enterococcus faecalis from necrotic root canals using multiplex PCR. J Oral Sci. 49: 221-227.
- Steenhoudt K. (1996). Bioreduction of Uranium by the Strain Enterococcus Faecalis. University of New Mexico, Bioremediation.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences