Molecular Docking Studies of Anthraquinone Derivatives on the Crystal Structure of Glycogen Synthase Kinase 3 Beta
Karthik Dhananjayan, Sumathy Arunachalam, Palanisamy Sivanandy
1Department of Pharmacology, Molecular Pharmacology and Drug Screening Division, Grace College of Pharmacy, Palakkad, Kerala, India;
2Department of Pharmaceutical Chemistry, Drug Design and Medicinal Chemistry Division, Grace College of Pharmacy, Palakkad, Kerala, India;
3Department of Pharmacy Practice, International Medical University, Kualalampur, Malaysia.
- Corresponding Author:
- Tel: 0491-2508537
Fax: 0491-2509393
E-mail: karthikdhanan@yahoo.com
Abstract
Glycogen synthase kinase 3β (GSK3β) is a serine/threonine kinase that has enzymatic activity regulated by many signaling pathways and by distinct multiproteincomplexes. Glycogen synthase kinase-3β (GSK-3β) has been investigated for its potential binding affinity with selective anthraquinones like aesculin, aesculetin, anthragallol, damnacanthal, lucidin, morindone, nordamnacanthal and Rubiadin. Insilico docking studies showed that almost every ligands with better binding affinity and a satisfactory inhibition constant. Among them damnacanthal, nordamnacanthal, Anthragallol and morindone showed lowest free energy of binding through interaction with potential areas of amino acids exposed at the active site crucial for enzyme inhibition. These ligands can be targeted towards the inhibition of Glycogen synthase kinase-3β (GSK-3β) involved in specific neoplastic disoders like prostate cancer, oral cancer etc.
Keywords
Anthraquinone, GSK3β, insilico docking, Neoplasia.
Introduction
Glycogen synthase kinase 3β(GSK3β) is a serine/threonine kinase that has enzymatic activity regulated by many signaling pathways and by distinct multiprotein complexes[1].Targeting GSK3β inhibition is of increased importance in the field of neoplasia. Being serine/threonine kinase, their impact lies on physiological roles and in diseases are based upon primed phosphorylated state of glycogen synthase [2].
Treating oral cancer through GSK3β inhibition on the basis of the inactivation of GSK3β may behave like an oncogene, and its gradual/sustained inactivation may promote oral cancer. Though most of the upstream and downstream targets and their expression status correlate with the understanding of GSK3β inactivation, real, direct assessment should be attempted. If the activated form of GSK3β is non-toxic to normal oral epithelial cells, as was found in animal models, then the manipulation of the activated GSK3β provides hope for treating oral cancer [3].
Glycogen Synthase Kinase-3 is involved in the phosphorylation and suppression of androgen receptor activity. Kinases can phosphorylate and regulate androgen receptor activity during prostate cancer progression. In particular, we showed that glycogen synthase kinase-3 phosphorylates the androgen receptor, thereby inhibiting androgen receptor-driven transcription. The glycogen synthase kinase-3inhibitor lithium chloride suppressed the glycogen synthase kinase-3-mediated phosphorylation of the androgen receptor, thereby enabling androgen receptor-driven transcription to occur [4].
Flavonoids showed invitro gsk3β inhibition [5]. Six anthraquinones (nordamnacanthal, alizarin-1-methyl ether, rubiadin, soranjidiol, lucidin-ω-methyl ether and morindone) isolated from the cell suspension culture of Morinda elliptica were assayed for antitumor promoting and antioxidant activities [6].
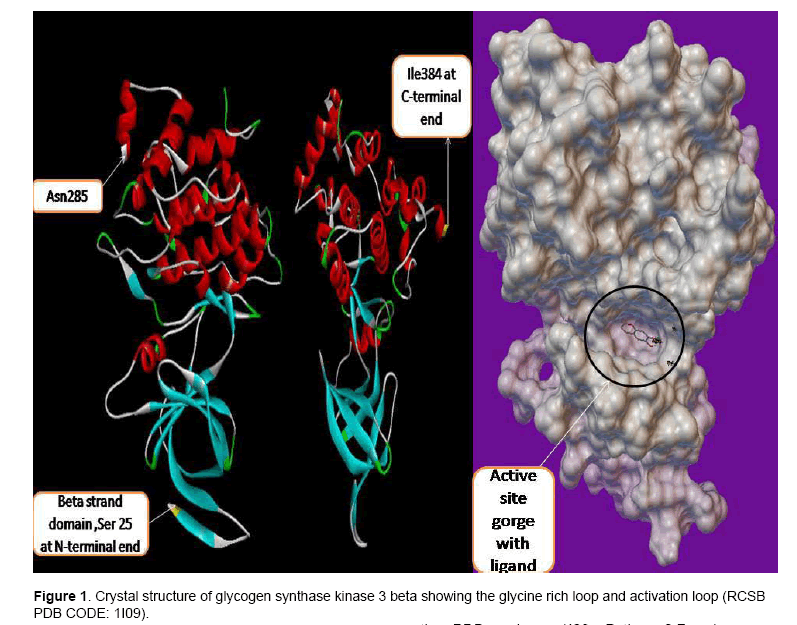
Figure 1 show the crystal structure of GSK3β. GSK3β posses typical two-domain kinase[7] fold with a β-strand domain (residues 25–138) at the N-terminal end and an α-helical domain at the C-terminal end (residues 139–343) . The ATP-binding site is at the interface of the α-helical and β-strand domain and is bordered by the glycine-rich loop and the hinge. The activation loop (residues 200–226) runs along the surface of the substrate binding groove. The C-terminal 39 residues (residues 344–382) are outside the core kinase fold and form a small domain that packs against the α-helical domain [8].
The anthraquinone derivatives were selected in order to analyse the binding affinity on the crystal structure of Glycogen synthase kinase 3 beta through computational docking study.
We have employed AutoDock 4.2 in order to evaluate the binding affinity. AutoDock 4.2 utilizes Monte Carlo simulated annealing and Lamarckian genetic algorithm (LGA) to create a set of possible conformations [9]. LGA is used as a global optimizer and energy minimization as a local search method. For the evaluation of possible orientations, AMBER force field model in conjunction with free energy scoring function is used. Coordinate files preparation, atomic affinities (AutoGrid) calculation. Semi empirical free energy force field is used to evaluate conformations during docking. The Ligand and protein stay in an unbound conformation. Then binding is evaluated in two steps by force field. Force field evaluates intramolecular energetics during the translation from their unbounded states to the conformation of both ligand and protein into the form of bound state [10].
2. Materials and Methods
The crystal structure of the investigational enzyme Glycogen Synthase kinase-3beta (gsk-3β) was downloaded from RCSB protein data bank bearing the PDB code – 1I09. Python 2.7 - language downloaded from www.python.com, cygwin- a data storage c:\program downloaded from www.cygwin.com, MGL (Molecular Graphics Laboratory) tools–AutoDock4.5 downloaded from www.scripps.edu, Chem sketch downloaded from www.acdlabs.com. Python 2.5 downloaded simultaneously during cygwin download, Accelry’s Discovery studio visualizer 3.1–downloaded from www.accelerys.com, Chem Office package- Chem 3D ultra- from www.cambridgesoft.com.
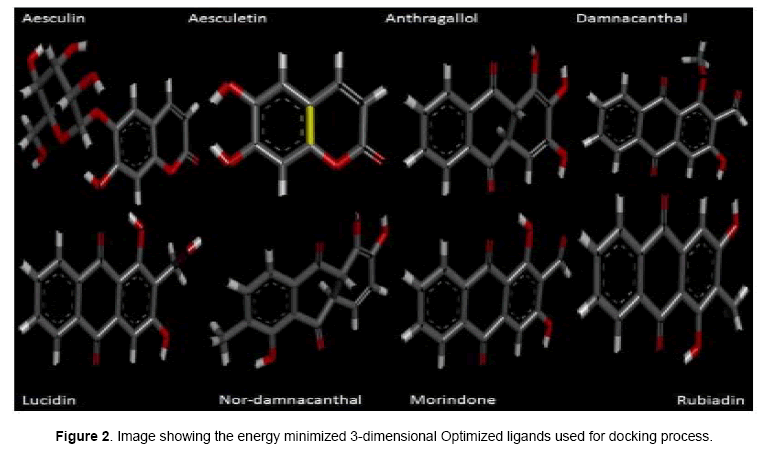
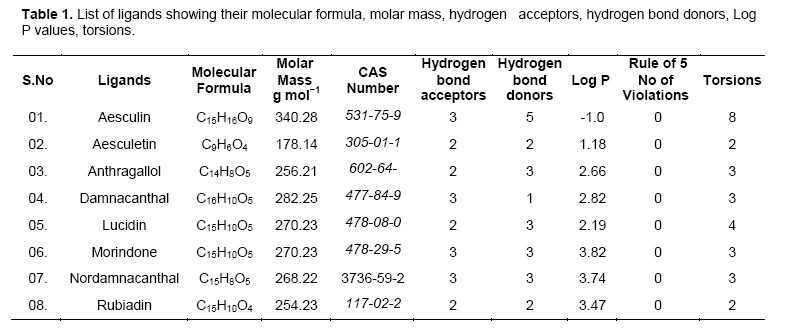
Investigational ligands have designed using Chem sketch. Designed ligands were converted to 3D structures and optimized for energy minimization using Chem 3D ultra. The optimized ligands were shown in Figure 2. The optimized ligands with their log P values, lipinski’s rule of 5 (no of violations), molar mass, molecular formula, and no of torsions are given in Table 1.
Molecular docking was performed in making enzyme molecule rigid and ligand to get flexible, in this way different conformation arises during each run and the best conformer fits with lowest binding energy (kcal/mol).
Rigid docking [10]was performed using the latest version of AutoDock 4.2.The enzyme molecule is loaded and stored as gsk3b.pdb after assigning hydrogen bonds and kollman charges. The investigative ligand was loaded and their torsions along their rotatable bonds are assigned and their file is saved as ligand.pdbqt. Grid menu is toggled, after loading enzyme.pdbqt the map files are selected directly with setting up grid points with 40 x40x40 dimensions for the searching of ligand within the active site of the enzyme molecule. This way the grid parameter files are created with setting up of map files directly
Followed by, setting up of docking parameter files with search parameter as genetic algorithm and docking parameter utilizing Lamarckian genetic algorithm. Then the docking process is carried out using cygwin interface and their results are viewed after final Lamarckian genetic algorithm gets completed successfully.
3. Results and Discussion
The free energy of binding elicited at the vicinity of active site by the ligand can be found from Table 2. Among the ligands, damnacanthal was found to possess lowest binding energy -7.23kcal/mol, with an estimated inhibition constant (kI) of 4.98uM. This free energy of binding was maintained in three conformations. The cluster RMSD values with respective to reference RMS values were given in Table 3.The chemically diverse nor-damnacanthal showed lowest binding energy of -6.94 kcal/mol with double the value of estimated inhibition constant, kI (8.25 μM) on comparing with damnacanthal. Anthragallol and morindone shared a similar binding affinity values (-6.68kcal/mol) and estimated inhibition constant, kI (12.7 μM) but their conformations at the active site was found to be varied. Rubiadin and lucidin has a slight deviation in energy creation at the actve site (-6.63 & -6.36 kcal/mol) but desired kI value of 13.92 μM 21.68 μM with respect to other ligands under study. Aesculetin and aesculin positioned with a binding affinity of -5.42 and -5.99 kcal/mol at the active site. This type free energy binding showed a higher kI values for aesculetin(105.78 μM) and aesculin (40.57 μM) when compared with other ligands.
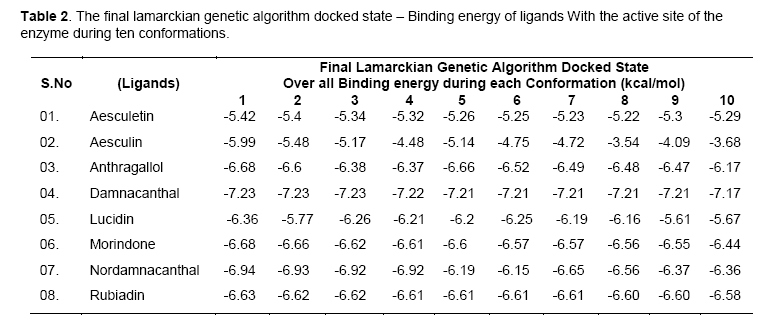
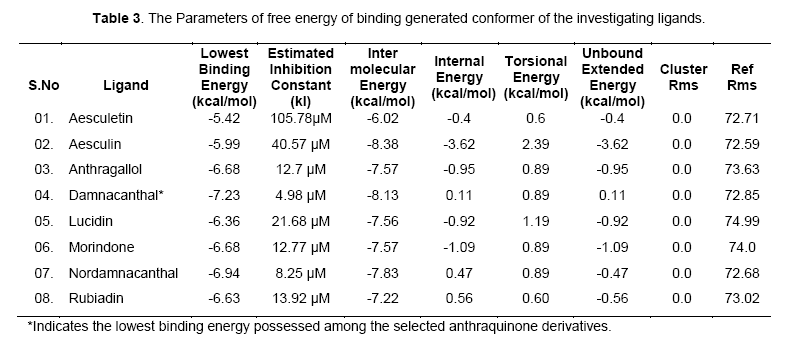
Free energy of binding (kcal/mol) created at the vicinity of an active site decides the potential interaction of a ligand and the protein (enzyme or a receptor) macromolecule. Rigid docking process carried out using Lamarckian genetic algorithm tends the ligands to be flexible at the receptor site, whereas the exposed amino acid residue stays as such rigid in their position. Maximum of ten poses generated by the ligands fixes up towards with its interaction energy. Lowest binding energy gives out the maximum potential interaction with a stable conformer [11]. Accordingly, investigational ligands like damnacanthal, nordamnacanthal, morindone, Rubiadin, lucidin involved in creation of lowest binding energies at the active site of targeted x-ray crystal structure of gsk3beta. The conformers generated at the active site depend upon the interaction energy of amino acid with the ligands. Among the amino acids exposed at the active site gorge, those involved in ligand interaction are Asn64, Ser66, Lys85, Val135, Thr138, Lys183, Asp200, Phe201. Analyzing the binding site and hydrogen bond length, complete information was obtained on the nature of interaction of functional group of ligand and amino acid residue of the enzyme at the phosphorylating site.
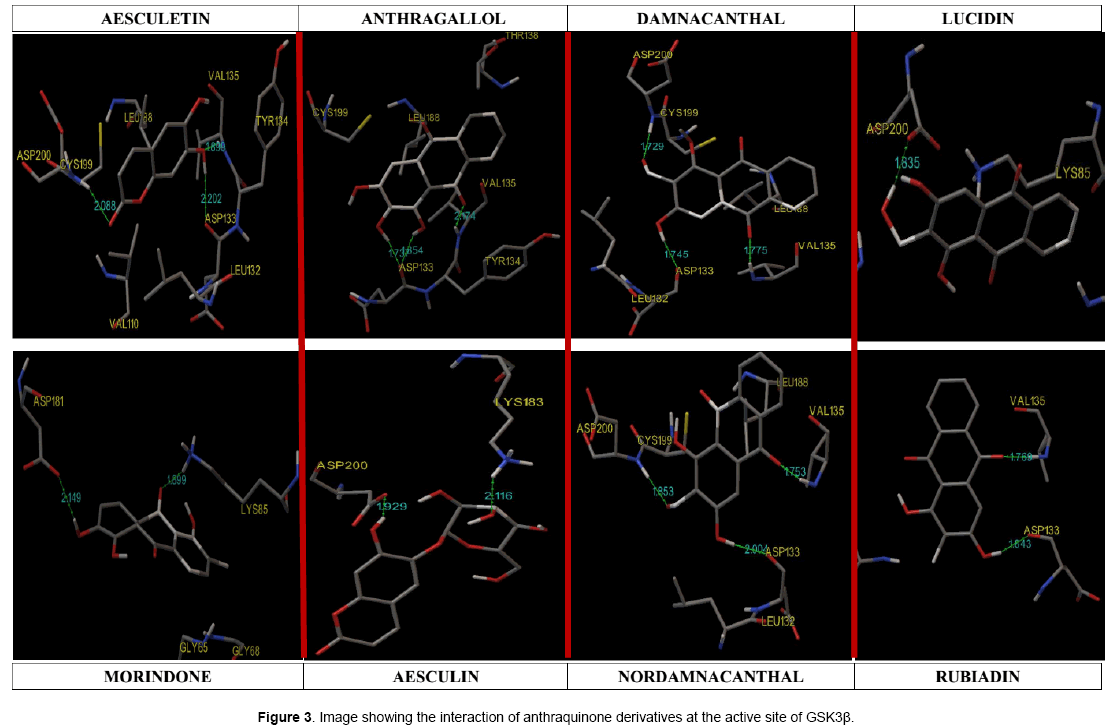
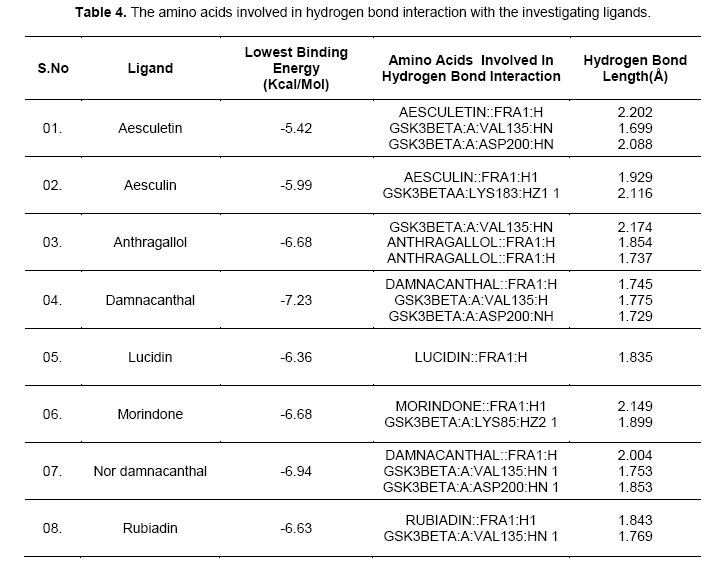
Hydrogen-bonds play a crucial role in determining the specificity of ligand binding. [12]. Table 4 shows the amino acids involved in hydrogen bonding with the ligands. It also lists out the length of hydrogen bonding in angstrom (Å). Figure 3 shows orientation of the ligands according to the free energy created at the active site with its hydrogen bond length. Damnacanthal is an anthraquinone, the lowest free energy of binding exhibited by damnacanthal and nor-damnacanthal interacts with Val135 (1.775 Å, 1.753 Å) and Asp200 (1.729 Å, 1.853Å) through hydrogen bonding. Next set of Anthraquinones, Morindone, Anthragallol and Rubiadin interacts at Lys85 (1.899 Å) and Val135 (2.174 Å,1.769 Å) through hydrogen bonding. Aesculetin and Aesculin with its lowest binding energy, Hbond with Val 135 (1.699 Å), Asp200 (2.088 Å) and Lys183 (2.116 Å). Ligands like thieno[2,3-b]pyrrolizinones interacted at the Val 135,Asp 200 and Lys183 with maximum affinity [13]. Binding affinity based estimated inhibition constant of aesculetin and aesculin was found to be higher than damnacanthal and nordamnacanthal. On analyzing the binding site interaction with respect to free energy of binding values anthraquinone derivatives containing hydroxyl groups and oxygen atom of anthraquinone nucleus interacts with amino acids like Val 135, Asp200, and Lys85 with maximum affinity.This is evident from the hydrogen bond length formed between the atom of ligand and the amino acid residue at the active site gorge.
Conclusion
Glycogen synthase kinase 3 beta has been investigated for its potential binding affinity with selective anthraquinone derivatives. It has been concluded, selective anthraquinone derivatives under our investigation showed maximum binding affinity with its estimated inhibition constant values at lower range. Binding site analysis on GSK3β reveals, the ligands occupies the dock positions, where phosphorylated glycogen synthase binds and carry out its interaction. Thus molecular signaling increasing the proliferation rate via cascading mechanism can be inhibited through these selective anthraquinone derivatives. Further it can be evaluated upon studying invitro gsk3beta inhibition and invivo models in bringing out a lead molecule targeting specific neoplasia like prostate cancer, oral cancer etc.
References
- Abdelvahab A. M., Abdalla M. H., (1995). The role of potassium fertilizer in nodulation and nitrogen fixation of faba beans (Vicia faba L.) plants under drought stress. J. Biol. Ferti Soils. 20: 147-150.
- A Kim L., and Kimmel A.R.(2000) GSK3, a master switch regulating cell-fate specification and tumorigenesis. Curr. Opin. Genet. Dev. 10: 508-514.
- Jia L., (2009) Glycogen synthase kinase 3β (GSK3β) in tumorigenesis and cancer chemotherapy. Cancer Letters, 273: 194–200.
- Rajakishore M. (2010) Glycogen Synthase Kinase 3 Beta: Can It Be A Target For Oral Cancer . Mol Cancer, 9: 144.
- Thomas RS., Jeri K., Funda, VL., et al. (2004.)Glycogen Synthase Kinase-3: Is Involved in the Phosphorylation and Suppression of Androgen Receptor Activity. J biol Chem, 279(18): 19191–19200.
- Jodee LJ., Sanjeewa GR., Felicia S., Mary A.S., Elvira G.M., (2011) Citrus Flavonoids Luteolin, Apigenin, and Quercetin Inhibit Glycogen Synthase Kinase-3bEnzymatic Activity by Lowering the Interaction Energy Within the Binding Cavity. J Med Food, 14(4): 325–333.
- Jasril NH., Laji Lim YM., Mohd AA., et al. (2003) Antitumor promoting and actioxidant activities of anthraquinones isolated from the cell suspension culture of Morinda elliptica. AsPac J. Mol. Biol. Biotechnol.. 11 (1): 3-7.
- Hanks SK., Quinn AM. (1991) Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol, 200: 38-62.
- Ernst TH., Joyce TC., Douglas AA., et al (2001) Structure of GSK3βreveals a primed phosphorylation mechanism. Nat Struct Biol, 8(7): 593-596.
- Garrett MM., David SG., Robert SH., et al.(1998) Automated Docking Using a Lamarckian Genetic Algorithm and an Empirical Binding Free Energy Function. Journal of Computational Chemistry, 19(14): 1639-1662 .
- Hay DV., Israel S, Michal H., Terrone, LR., Joel LS. (2010) Acetyl cholinesterase: From 3D structure to function Chem-Biol Interact; 187: 10–22.
- Bjørn OB., Fredrik Ö., Martin A., Isabella F., Victor BL., Johan A., et al. (2003) Free Energy Calculations and Ligand Binding. Adv. in Prot Chem ,66: 123-158.
- Wade RC., Goodford PJ. (1989) The role of hydrogen-bonds in drug binding. Prog Clin Biol Res, 289: 433-44.
- Lodie L., Ronan B., Jana OS, et al. (2005) 3D-QSAR And Docking Studies Of Selective GSK-3β Inhibitors. Comparison With A Thieno[2,3-B]Pyrrolizinone Derivative, A New Potential Lead For GSK-3β Ligands. J. Chem. Inf. Model., 45 (3): 708–715.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences