First Isolation of Leptospira borgpetersenii from Fetuses of Wild Boars (Sus scrofa)
Bibiana Brihuega, Sylvia Grune Loffler, Luis Samartino, Graciela Romero, Carmelo Auteri, Mara Martinez
1Institute of Pathobiology, Laboratory of Leptospirosis, National Institute of Agricultural Technology, Hurlingham, Buenos Aires, Argentina
2National Research Council of Argentina (CONICET), Buenos Aires, Argentina.
- Corresponding Author:
- Sylvia Grune Loffler
Institute of Pathobiology, Laboratory of Leptospirosis
National Institute of Agricultural Technology, Hurlingham, Buenos Aires
Argentina; National Research Council of Argentina (CONICET)
Buenos Aires, Argentina
Tel: Tel: 0054-11-46211289
E-mail: grune.sylvia@inta.gob.ar
Received date: January 03, 2017; Accepted date: January 30, 2017; Published date: February 07, 2017
Citation: Brihuega B, Loffler SG, Samartino L, et al. First Isolation of Leptospira borgpetersenii from Fetuses of Wild Boars (Sus scrofa). Electronic J Biol, 13:1
Abstract
Leptospirosis is a worldwide zoonose and is endemic in many countries. This zoonosis is maintained in nature by chronic renal infection of carrier animals, being rodents and other small mammals the most important reservoirs. Also, significant sources of human infection are domestic animals, such as livestock and dogs. The wild boar (Sus scrofa) is an introduced species from Europe which is widespread in America and particularly in the Center and South of Argentina. This species is highly valued by hunters in the region and their meat is consumed more often. In many countries of the world (Europe, USA and Australia) studies of seroprevalence have been carried out, the seroreactivity does not mean that the wild boar has clinical symptoms of leptospirosis or that wild boars are maintenance host of this pathogen. However, these animals have been in contact to leptospiras in their environment in the past. This study has the objective of molecular characterization of pathogenic Leptospira sp. isolated strain obtained from fetuses of wild boars from Patagonia Argentina. Four fetuses were aborted from a wild boar (Sus scrofa) in Patagonia, the southern region of Argentina. Necropsy was performed of the 4 fetuses and samples of kidneys, liver, spleen and lung were cultivated in Fletcher and EMJH mediums. Also, samples of liver, spleen, lung and kidney of fetuses were processed and analyzed by direct immunofluorescence. Multiple Locus Variable number tandems repeats Analysis (MLVA) was used to characterize the isolated strain. The genetic profile of the isolated pathogenic Leptospira sp. strain was identical to the profile of Leptospira borgpetersenii serovar Castellonis Castellon 3. To the best of our knowledge this is the first isolation of Leptospira spp. from a wild boar fetus.
Keywords
Leptospira spp.; Wild boar; Genotype; Patagonia; Argentina.
Introduction
Leptospirosis is the most widespread zoonotic disease in the world. It is caused by pathogenic Leptospira spp. spirochetes, which find an ideal niche for transmission in subtropical and tropical regions. There are no available data on the incidence of leptospirosis in several countries, however in addition, the disease is frequently not recognized and, consequently, severely neglected [1]. Pathogenic Leptospira spp. strains mainly infect mammals, but they can also be found in reptiles and amphibians [2,3]. This zoonosis is maintained in nature by chronic renal infection of carrier animals, being rodents and other small mammals the most important reservoirs. In addition, significant sources of human infection are livestock and domestic animals, such as dogs. In Argentina this disease is endemic and no epidemiological control or prevention programs have been yet implemented. Until November 2016, an estimate of 3206 human cases was notified to the National Health Ministry of Argentina (National Health Ministry, 2016) [4].
Ellis [5] established domestic pigs as maintenance hosts for the serogroup Australis and in Jansen et al., for serogroup Pomona. On the other hand wild boars are categorized as accidental hosts for serovar Grippothyphosa in Treml et al. [6] and in Jansen et al. [7] for serogroup Pomona and Bratislava. In Slavica et al. [8] a serological study in wild boars in Croatia was carried out using MAT, in this study the most frequent serovars were Australis (33,3%), Pomona (21,8%) and Tarassovi (14,3%) from a total of 351 serum samples, the highest titre obtained was 1/3200 for serovar Pomona. In a recent serological study of wild boars (Sus scrofa) in Poland 3621 blood samples from wild boars were analyzed for following Leptospira spp. serovars/serogroup [9]: Icterohaemorrhagie, Grippothyphosa, Sejore, Tarassovi, Pomona, Canicola, Bratislava, Autumnalis, Hardjo and Ballum. The serogroup Ballum did not gave titers in the serum samples tested. Serovars Pomona and Sejore are reported as the most common in domestic pigs in Poland; however in Zmudzki et al. [9] these serovars were infrequent. In the same study, increase of free living wild boar population in Europe is mentioned in urban and suburban areas and the threat of possible dissemination of pathogenic Leptospiral strains is concerning. The necessity for population control also constitutes a risk for hunters to become infected by Leptospira spp. [10,11]. Also the geographic expansion of humans into not urbanized areas is causing that wildlife animals have to live closely with the inhabitants of these towns and cities. In 2006, a human case of leptospirosis in Berlin was linked to the possible infection by freshwater contaminated with wild boar urine [11]. In a review of wild boars as sources of infections in humans and livestock (Meng et al. [12]), only three studies are listed for positive seroreactivity: Germany (Jansen et al. [7]), California (USA) (Clark et al. [13]), and Italy (Ebani et al. [14]). Also in Oklahoma (USA) Saliki et al. [15] was also found seroreactivity in wild boars (Sus scrofa). In Jansen et al. [7] serum samples of wild boars of urban areas of Berlin were tested by MAT, serovar Pomona and Bratislava were the most frequent, these results and the demonstration that leptospires were found by silver staining in kidney samples of wild boars showed that these animals are maintenance hosts also in urban areas. In Slovenia Vengust et al. [16] in a seroprevalence study of Leptospira spp. in 437 wild boar serum samples, found that the prevalence of antibodies of this study was higher (45.5%) than the prevalence determinate in previous studies: Italy (6%; Ebani et al. [14]), Czech Republic (16.9%; Treml et al. [6]), Germany (24%; Schönberg et al. [17]), Poland (25.2%; Krawczyk [18]), Croatia (26%; Cvetnic et al. [19]), Austria (30%; Deutz et al. [20]) and Australia (20%; Mason et al. [21]).
In Brazil a serological screening using MAT was carried out in a captive population of wild boars for human consumption. In this study 63 animals were positive to Leptospira spp. the best represented serovars was: Hardjo, Copenhageni and Pomona [22].
To the best of our knowledge this is the first isolation of L. borgpetersenii from fetuses of wild boars (Sus scrofa).
Materials and Methods
Isolation of Leptospira spp
Necropsy was performed of the 4 fetuses (Figure 1) using the protocol described by Faine et al. [23] and samples of the kidneys, liver, spleen and lung were cultivated in Fletcher and EMJH media. The cultures were incubated at 28°C until development was determinate, every 15 days the cultures were observed under dark field microscopy.
Immunofluorescence
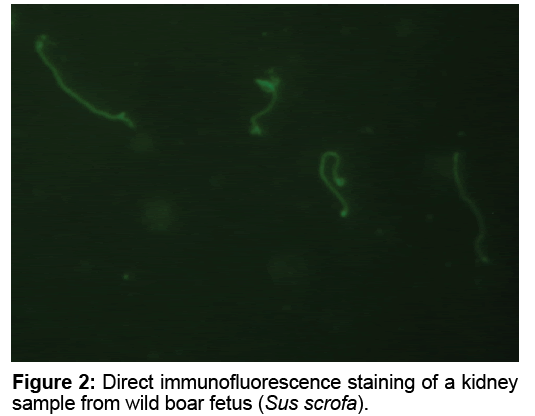
Direct immunofluorescence staining of liver, spleen, lung and kidney samples were performed with a multivalent FA, LEP-FAC (Seasinglab, Argentina) conjugate specific for Leptospira spp. (Figure 2)
Molecular characterization
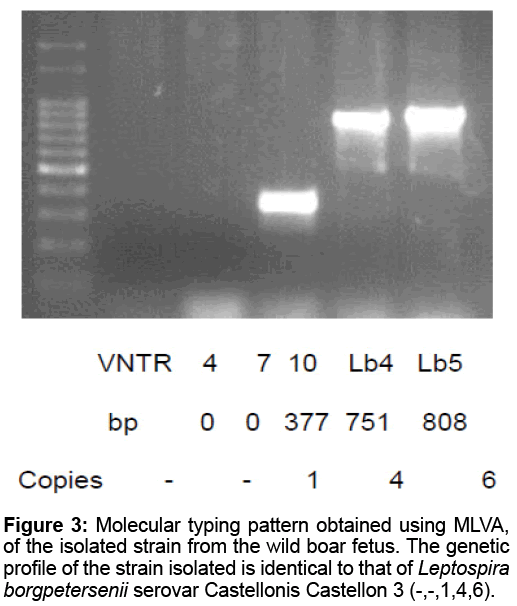
The reference strains and the strain isolated were grown in Fletcher media (Difco Laboratories) at 28ºC. For the DNA templates the protocol used with the resin Chelex-100 (Bio Rad) was used. Multiple-Locus Variable-number tandem repeats Analysis (MLVA) was used to characterize the isolated strain with 1 set of oligonucleotides specific for the pathogenic strains of L. interrogans, L. kirschneri and L. borgpetersenii, following loci were used, VNTRS: 4, 7, 10, Lb4 and Lb5 [24]. The final volume (50 μl) of each reaction mixture contained PCR Buffer (20 mMTris- HCL,pH 8.4;50 mM KCL), 200 μM deoxinucleoside triphosphates, 2 μM each corresponding primer, 2 mM MgCl2, 1.25 U Taq DNA polymerase (Invitrogen) and 5 μl DNA template. PCRs were carried out in a Thermo Scientific PxE 0,2 Thermal Cycler as follows: 94ºC for 5 min, followed by 35 cycles of denaturalization at 94ºC for 30 s, annealing at 55ºC for 30 s and extension at 72ºC for 90 s, with a final cycle at 72ºC during 10 min. Amplified samples (15 μl) were revealed by electrophoresis in a 2% agarose gel in buffer TAE (40 mM Tris-acetate, 1mM EDTA) with 0,2 ul μg/ml of ethidium bromide at 100 V for 50 min. Amplified DNA bands were visualized upon UV light exposure (Uvi Tec transiluminator BTS-20.M). Amplicon sizes were estimated using CienMarker (Biodynamics) and the GelAnalyzer 2010a program. To calculate the repeat copy number the following formula was used: Number of repeat (bp)=[Fragment size (bp)-Flanking regions (bp)]/Repeat size (bp). Repeat copy numbers were rounded down to the closest whole numbers. If the copy number was less than one it was rounded to zero.
Results
Direct Immunofluorescence was positive in liver, spleen and kidney of fetuses. One Leptospira spp. strain was isolated from kidneys and liver in Fletcher medium after 47 days of incubation at 28ºC. The genotyping of the isolation is coincident with the genetic profile of Leptospira borgpetersenii serovar Castellonis Castellon 3 (-,-,1,4,6) of serogroup Ballum (Figure 3).
Discussion and Conclusion
To the best of our knowledge, this is the first isolated strain of Leptospira borgpetersenii isolated from wild boars in South America and the first isolated strain from aborted wild boar fetuses. This finding suggests that wild boars are susceptible to L. borgpetersenii serovar Castellonis Castellon 3 of serogroup Ballum. This genotype was previously characterized in mice populations in Argentina [25]. The same species L. borgpetersenii was isolated in water samples in an urban river in Argentina by Francoise et al. [26] and has been isolated in clinical cases of domestic animals by Grune et al. [27] and in humans by Goncalves et al. [28]. Rossetti et al. [29] isolated L. interrogans from domestic pigs in Argentina. This species has been isolated also from free-living opossums in Brazil [30]. Several serosurvey studies carried out in wild boars (Europe, USA and Australia) show the exposure in the environment and ecological niches where wild boar populations live, to pathogenic Leptospiral strains in their past, but in this study an isolated strain from an aborted fetus of Sus scrofa could be cultured and molecular characterized.
Acknowledgement
This research was financed by Proyecto Nacional INTA, AESA 202821.
References
- Hartskeerl RA, Collares-Pereira M, Ellis WA. (2011). Emergence, control and re-emerging leptospirosis: Dynamics of infection in the changing world. Clin Microbiol Infect. 17: 494-501.
- Levett PN. (2001). Leptospirosis. Clin Microbiol Rev. 14: 296-326.
- Adler B. Peña Moctezuma A. (2010). Leptospira and leptospirosis. Vet Microbiol. 27: 287-296.
- National Health Ministry. (2016). Ministerio de Salud. Presidencia de la Nación. Boletín Integrado de Vigilancia. Dirección Nacional de Epidemiología y Análisis de la Situación de Salud. SE51: 96.
- Ellis WA. (2015). Animal Leptospriosis. In: Leptospira and Leptospirosis. Ed. Ben Adler. Springer-Verlag Berlin Heidelberg, Germany. Curr Top Microbiol Immunol. 387: 1.
- Treml F, Pikula J, Holesovska Z. (2003). Prevalence of antibodies against leptospires in the wild boar (Sus scrofa L., 1758). Vet Med Czech. 48: 66–70.
- Jansen A, Luge E, Guerra B, et al. (2007). Leptospirosis in urban wild boars, Berlin, Germany. Emerging Infectious Diseases. 13: 739-742.
- Slavica A, Cvtnic Z, Konjevic D, et al. (2010). Detection of Leptospira spp. serovars in wild boars (Sus scrofa) from continental Croatia. Veterinarski Archiv. 80: 247-257.
- Zmudzki J, Jablonski A, Nowak A, et al. (2016). First overall report of Leptospira infections in wild boars in Poland. Acta Vet Scand. 58: 3.
- Richard S, Oppliger A. (2015). Zoonotic occupational diseases in forestry workers—Lyme borreliosis, tularemia and leptospirosis in Europe. Ann Agric Environ Med. 22: 43–50.
- Jansen A, Nöckler K, Schönberg A, et al. (2006) Wild boars as possible source of hemorrhagic leptospirosis in Berlin, Germany. Eur J Clin Microbiol Infect Dis. 25: 544–546.
- Meng XJ, Lindsay DS, Sriranganathan N. (2009). Wild boars as sources for infectious diseases in livestock and humans. Phil Trans R Soc B. 364: 2697-2707.
- Clark R, Jessup DA, Hird D, et al. (1983). Serologic survey of California wild hogs for antibodies against selected zoonotic disease agents. J Am Vet Med Assoc. 183: 1248-1251.
- Ebani VV, Cerri D, Poli A, et al. (2003). Prevalence of Leptospira and Brucella antibodies in wild boars (Sus scrofa) in Tuscany, Italy. J Wildl Dis. 39: 718–722.
- Saliki JT, Rodgers SJ, Eskew G. Serosurvey of selected viral and bacterial diseases in wild swine from Oklahoma. 1998. J Wildlife Dis. 34: 834–838.
- Vengust G, Lindtner-Knific R, Zele D, et al. (2008). Leptospira antibodies in wild boars (Sus scrofa) in Slovenia. European Journal of Wildlife Research. 54: 749–752
- Schönberg A, Lutz W, Kampe U. Investigation of serum samples of wild boar (Sus scrofa L-1758) for Leptospirosis. Z Jagdwiss. 45: 262–265
- Krawczyk M. (2000). Serological studies on leptospirosis in wild boar. Med Weter. 56: 440–443
- Cvetnic Z, Margaletic J, Toncic J, et al. (2003). A serological survey and isolation of leptospires from small rodents and wild boars in the Republic of Croatia. Vet Med Czech. 11: 321–329.
- Deutz A, Fuchs K, Schuller W, et al. (2002). Studies on the seroprevalence of antibodies against Leptospira interrogans in hunters and wild boar from south–eastern Austria. Z Jagdwiss. 48: 60–65
- Mason RJ, Fleming PJ, Smythe LD, et al. (1998). Leptospira interrogans antibodies in feral pigs from New South Wales. J Wildlife Dis. 34: 738–743.
- Fornazari F, Camossi LG, Silva RC, et al. (2011). Leptospiral antibodies in wild boars (Sus scrofa) bred in Brazil. J Venom Anim Toxins incl Trop Dis. 17: 94-97.
- Faine S, Adler B, Bolin C, Perolat P. (1999). Leptospire and leptospirosis (2nd edn). MediSci; Melbourne, Australia. 272.
- Salaün L, Mérien F, Gurianova S, Baranton G, Picardeau M. (2006). Application of multilocus variable-number tandem-repeat analysis for molecular typing of the agent of leptospirosis. J Clin Microbiol. 44: 3954-3962.
- Grune Loffler S, Pavan ME, Vanasco B, et al. (2014). Genotypes of pathogenic Leptospira spp. isolated from rodents in Argentina. Mem Inst Oswaldo Cruz. Rio de Janeiro. 1-5.
- François S, Brihuega B, Grune S, et al. (2013). Aislamiento de Leptospira borgpetersenii de fuentes de agua en Argentina. Revista Cubana de Medicina Tropical. Nr. 2.
- Grune Loffler S, Samartino L, Brihuega B. (2017). Insights into genetic distances of isolated strains of pathogenic Leptospira spp. from humans, animals and environment using multiple locus variable-number tandem repeat analysis (MLVA) genotypes. J Zoonotic Dis Public Health. 1:1.
- Goncalves A, Paiva C, Melo-Mota F, et al. (2010). First isolation of human Leptospira strains, azores, Portugal. Int J Infect Dis. 14: 148-153.
- Rossetti C, Brihuega B, Auteri C, et al. (2002). Leptospirosis porcina en la República Argentina: Encuesta serológica y primera comunicación del aislamiento de una cepa de Leptospira interrogans (sg. Icterohaemorragiae) de una cerda abortada. Rev Vet Arg. 17: 578-582.
- Jorge S, Hartleben CP, Seixas FK, et al. (2012). Leptospira borgpetersenii from free-living white-eared opossum (Didelphis albiventris): First isolation in Brazil. Acta Trop.124: 147-151.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences