The Role of CpG24 Hyper-Methylation on the Down-Regulation of Rap1Gap in Papillary Thyroid Cancer
Bita Faam, Ata Ghadiri, Mehdi Totonchi, Atieh Amouzegar, Ahmad Fanaei, Mohammad Ali Ghaffari*
Published Date: 2018-05-05Bita Faam1, Ata Ghadiri2, Mehdi Totonchi3, Atieh Amouzegar4, Ahmad Fanaei5, Mohammad Ali Ghaffari1,6*
1Cellular and Molecular Research Center, Joundishapour University of Medical Sciences, Ahvaz, Iran;
2Department of Immunology, Faculty of Medicine, Joundishapour University of Medical Sciences, Ahvaz, Iran;
3Department of Endocrinology and Female Infertility, Reproductive Biomedicine Research Center, Royan Institute for Reproductive Biomedicine, ACECR, Tehran, Iran;
4Endocrine Research Center, Research Institute for Endocrine sciences, Shahid Beheshti University of Medical Sciences, Tehran, Iran;
5Erfan Grant Hospital, Tehran, Iran;
6Department of Biochemistry, School of Medical, Joundishapour University of Medical Sciences, Ahvaz, Iran.
Received date: March 27, 2018; Accepted date: April 17, 2018; Published date: May 05, 2018
Citation: Faam B, Ghadiri A, Totonchi M, et al. The Role of CpG24 Hyper-Methylation on the Down-Regulation of Rap1Gap in Papillary Thyroid Cancer. Electronic J Biol, 14:2
Abstract
Background: Thyroid tumors may arise following hypermethylation in the regulatory region of suppressor genes such as Rap1Gap. In this study, we aimed to investigate the gene expression of Rap1Gap and DNA methylation pattern of CpG24 in Iranian patients with papillary thyroid cancer (PTC).
Materials and methods: The study was done on 132 individuals who had undergone thyroidectomy. Rap1Gap mRNA expression was examined by an optimized two-step real-time quantitative PCR assay. DNA methylation pattern of CpG24 was examined using the methylation-specific PCR (MSP). B-CPAP and SW1736 cell lines were treated with 5-aza-2,-deoxycytidine (5-Aza) in an attempt to demethylate the CpG24 in the regulatory region of the target gene.
Results: The results of qRT-PCR indicated a no significant decrease of Rap1Gap gene expression level in PTC individuals compared with control group (fold change: 0.98; p=0.99). CpG24 was hyper-methylated in 82.3% of individuals with PTC, in 79.1% of benign nodules and only in 14% of normal thyroid tissues (p<0.001). The modest CpG24 de-methylation was achieved by 15 μM of 5-Aza in both cell lines.
Conclusion: It is concluded that aberrant DNA methylation in the CpG24 region, independent of Rap1Gap gene expression, could be determined as an epigenetic marker for papillary thyroid cancer.
Keywords
Epigenetic; DNA methylation; Papillary thyroid cancer; Tumor suppresser gene; Gene expression
Introduction
Thyroid cancer is the most prevalent endocrine malignancy and its incidence is increasing worldwide [1]. Papillary thyroid cancer (PTC) accounts for approximately 80% of all thyroid cancer [2]. Genetic and epigenetic modifications are involved in initiation and progression of thyroid cancer, of which mutations leading to the mitogen-activated protein kinase (MAPK) and phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) signaling pathways activation are crucial [3,4]. Other genetic and epigenetic factors which contribute to the malignant prognosis of PTC are insufficiently defined. Understanding these molecular alterations and their mechanisms may result in the development of novel molecular prognostic and therapeutic strategies [5-7]. DNA hyper-methylation is one of the epigenetic regulatory mechanisms which may silence the target gene without affecting the DNA sequence.
Rap1Gap gene in the 1p36 region encodes a type of GTPase-activating protein (GAP) that down-regulates the activity of Ras-related Rap1 protein. The product of Rap1Gap gene promotes the hydrolysis of bound GTP and returns Rap1 to the inactive state. RAS superfamily proteins, such as Rap1, play key roles in receptor-linked signaling pathways that control cell growth and differentiation. Rap1 plays the role in diverse processes such as cell proliferation, differentiation, and embryogenesis [8]. It is reported that Rap1Gap is a tumor suppressor gene which is down-regulated in various cancers such as squamous cell carcinoma, renal cell carcinoma, melanoma and thyroid cancer [9-12]. Rap1Gap expression and its activity are mostly regulated at transcriptional and post-transcriptional levels. Tumors may arise following hyper-methylation in the regulatory region of Rap1Gap gene [13]. Understanding the pattern of tumor suppressor gene down-regulation and underlying molecular mechanisms, in thyroid cancer, may provide a significant clinical impact. In this study, we aimed to investigate the gene expression of Rap1Gap and DNA methylation pattern of CpG24 region in Iranian patients with papillary thyroid cancer.
Materials and Methods
Patients and tissue specimens
A total of 132 thyroid samples, diagnosed for normal thyroid tissue (n=50), benign nodule (n=48) and PTC (n=34), were obtained at the time of surgical resection between 2013 and 2015 from the Erfan Grand Hospital in Tehran, Hospital of Emam Khomeini and Arvand in Ahvaz. Normal samples were reviewed to ensure that no microcarcinoma or lymphocytic infiltrate was present in the tissue. The size of PTC tumors was obtained 0.5 to 9 cm, as well as there were demonstrated 8 ones had lymph node involvement. Tissue samples were immediately snapped frozen in liquid nitrogen and stored at -80°C. Data of patients were collected after providing written informed consent to the committee of ethical research of vice chancellery for research and technology development Ahvaz Joundishapour University of Medical Sciences (code: ajums. REC.1393.151). Pathological diagnosis was independently confirmed by two experienced pathologists.
Cell lines
B-CPAP and SW1736 cell lines were used from the tumor tissue of old women with metastasizing papillary thyroid cancer and anaplastic thyroid cancer, respectively. These cell lines were provided by Pasteur Institute of Iran and Iranian Biological Resource Center, respectively. Cells were cultured in the RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 u/ml penicillin and 100 u/ml Streptomycin at 37°C in a humidified atmosphere containing 5% CO2.
mRNA preparation
Total RNA of tissues and cell lines were extracted using RiboEx (Trizol) (Gene All Bldg, Seoul Korea) according to the manufacturer's instructions. Single-stranded cDNA was synthesized from 1 μg of total RNA in a 20 μl reaction and oligo (dt) primer (synthesized by Takara Bio Inc., Shiga, Japan). Quality of mRNA and cDNA were determined using the NanoDrop spectrometer (NB-1000, USA).
Gene expression
Rap1Gap mRNA expression was examined by an optimized two-step real-time quantitative PCR assay. The primers sequences were checked with qprimer depot database (Wenwu Cui, PhD Laboratory of Receptor Biology and Gene Expression, E-mail: byunis@mail.nih.gov) (Table 1). PCR amplification mixture (20 μl) contained SYBR Green PCR master mix (10 μl, 2x, applied biosystem), 0.5 μl of each forward and reverse primers and template cDNA (70 ng total RNA equivalent). Reactions were run on an ABI step one 48 (ABI amplicone). The cycling conditions were as follow: 15 min at 95°C, 40 cycles at 95°C for 30 s and 63°C for 60 s. All assays were performed in duplicate. After PCR amplification, a melting curve was generated for every PCR product to ensure the specificity of the reaction. Data were analyzed according to the relative standard curve method, in which the transcription levels were normalized by the stably expressed reference gene GAPDH (glyceraldehydo-3-phosphate dehydrogenase).
| Target | Primer sequences | AT (°C) | Product size (bp) |
|---|---|---|---|
| GAPDH | F: 5´-ctcatttcctggtatgacaacga-3´ | 61.1 | 121 |
| R: 5´-cttcctcttgtgctcttgct-3´ | 58.4 | ||
| Rap1Gap | F: 5´-acgagcatgtcatcagcaat-3´ | 56.4 | 138 |
| R: 5´-ccttctggccaagaaattca-3´ | 56.4 | ||
| Rap1Gap, MSP (U) | F: 5´-tagagataaagtttaagagttgtga-3´ | 54.8 | 220 |
| R: 5´-ttctaaatcaaataaaaacatcaaa-3´ | 49.9 | ||
| Rap1Gap, MSP (M) | F: 5´-tagagataaagtttaagagtcgcga-3´ | 58.1 | 220 |
| R: 5´-ttctaaatcaaataaaaacgtcgaa-3´ | 53.1 |
MSP: Methylation Specific PCR; M: Methylated; U: Un-Methylated; F: Forward; R: Reverse; AT: Annealing Temperature; bp: Base Pair
Table 1: The primer sequences used in qPCR amplification and methylation specific PCR.
Methylation-specific PCR (MSP)
Genomic DNA was extracted from thyroid tissues and cell lines using salting out method. DNA quality was determined using the Nano Drop spectrometer (NB-1000, USA). The amount of 0.35 μg of DNA was sodium bisulfate converted using the Ez DNA Methylation-Direct TM kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s protocol, during which methylated cytosine is protected and un-methylated cytosine is converted to uracil. Subsequently, 200 ng of treated DNA was used as a template in the Methylation Specific PCR (MSP) reaction. The primers were designed to attach to the CpG24 region of Rap1Gap gene. MSP was performed with methylationspecific primers sequences are shown in (Table 1). Each PCR reaction was performed with a total volume of 15 μl, which contained 7.5 μl of Hot-Star Taq Master Mix (Ampliqon), 200 ng of bisulfite-treated DNA template and 0.01 μM of each primer pair. The annealing temperature of PCR amplification was 60°C for methylated primer pair and 56°C for un-methylated primer pair. The reaction mixture was incubated at 94°C for 3 min, followed by 35 cycles of 94°C for 60 s, 60 or 56°C for 60 s, 72°C for 60 s and a final extension at 72°C for 10 min. Each reaction was tested with untreated DNA to ensure a lack of amplification and two controls were included to ensure specificity. After electrophoresis on 2% agarose gel, MSP products were analyzed.
5-aza-2,-deoxycytidine treatment of cell lines
B-CPAP and SW1736 cell lines were grown for 24 h before treatment as the control cells, then they were treated with 5-aza-2,-deoxycytidine (5,-aza; sigma) in an attempt to demethylate the CpG24 in the regulatory region of the target gene. Cells were cultured in the RPMI1640 medium and then 5-aza was added with concentrations of 5 μM, 15 μM and 25 μM during 72 h treatment before the extraction of DNA and RNA.
Statistical analysis
Quantitative RT-PCR results were assessed using REST 2009 software (Qiagene, Hilden. Germany). The chi-square test was applied to analyze the frequency of methylation in thyroid tissues; this statistical test was also used to assess the correlation of CpG24 methylation and lymph node involvement. The correlation of Rap1Gap gene expression with methylation status and also the correlation of tumor size with methylation status of CpG24 in papillary thyroid cancer group were evaluated by Spearman correlation test. All statistical analysis was done with SPSS software, version 20 (SPSS, Inc. Chicago, IL, USA). P ≤ 0.05 was considered to indicate the statistically significant difference.
Results
This study was done on 132 individuals (25 (19%) males and 107 (81%) females) who had undergone thyroidectomy. From among this population, 34 subjects had PTC, 48 subjects had benign nodules, and 50 individuals had healthy thyroid tissue (Table 2). The mean age of participants was 47.4 ± 12.1 years. The lowest age was significantly observed in the PTC group in compare with benign nodule and control groups (PTC: 40.0 ± 8.6 years; benign: 49.4 ± 12.8 years; control group: 48.7 ± 10.5 years; p=0.07). The PTC group contained four males and 30 females. Total studied population were overweight (25 ≤ BMI<30 kg/m2); but there was obtained a no significant difference at BMI of benign and PTC groups in compare with control group (benign: 28.9 ± 6.6 kg/m2; PTC: 26.6 ± 0.57 kg/m2; control: 28.2 ± 4.9 kg/m2; p=0.661). The mean size of tumor was 4.75 ± 1.91 cm in PTC population; this measurement was 6.4 ± 2.8 cm among CpG24 hypermethylated samples and 3.5 ± 1.2 cm among CpG24 unmethylated samples. Methylation pattern of CpG24 was significantly correlated with tumor size (p=0.036).
| Variables | Values |
|---|---|
| Sex (%) | |
| Male | 25 (19) |
| Female | 107 (81) |
| Pathology (%) | |
| Control | 50 (37.9) |
| Benign | 48 (36.4) |
| PTC | 34 (25.7) |
| Methylation status (%) | |
| UU | 59 (44.7) |
| UM | 71 (53.7) |
| MM | 2 (1.5) |
PTC: Papillary Thyroid Cancer; UU: Un-methylated Pattern; UM: Hetero-methylated Pattern; MM: Methylated Pattern
Table 2: A descriptive of anthropometrics data and CpG24 methylation status in patients.
From among PTC population, 8 (23.5%) ones had lymph node involvement and the Rap1Gap methylation was correlated with this characteristic (p=0.04).
Rap1Gap gene expression
The results of qRT-PCR indicated a no significant decrease of Rap1Gap gene expression level in PTC individuals compared with control group (fold change: 0.98; p=0.99). There was also obtained a no significant diminish in the expression level of Rap1Gap gene in benign group when they were compared to control group (fold change: 0.85; p=0.87). Finally, our results no significantly showed a reduction in Rap1Gap gene expression level of the papillary thyroid cancer samples compared with benign group (fold change: 0.96; P=0.83) (Table 3). There was demonstrated similar findings in thyroid cancer cell lines before treatment with 5-Aza. It was showed a no significant down-regulation of Rap1Gap in BCPAP cells (fold change: 0.83; p=0.95) and in the SW1736 cell line compared with control thyroid tissue (fold change: 0.20; p=0.79).
| Fold change | 95% CI | Average value of 95% CI | P-value | |
|---|---|---|---|---|
| Thyroid tissues | ||||
| PTC/Control | 0.99 | 0.02-6.45 | 3.22 | 0.99 |
| Benign/Control | 0.855 | 0.001-9.55 | 4.77 | 0.873 |
| PTC/Benign | 0.98 | 0.05-21.6 | 10.8 | 0.832 |
| BCPAP cell line | ||||
| [5]/[0]* | 0.841 | 0.69-1.02 | 1.18 | 0.517 |
| [15]/[0] | 0.927 | 0.79-1.12 | 0.95 | 0.354 |
| [25]/[0] | 0.888 | 0.79-0.99 | 1.28 | <0.001 |
| SW1736 cell line | ||||
| [5]/[0]* | 4.44 | 3.46-5.72 | 4.58 | <0.001 |
| [15]/[0] | 9.8 | 8.64-11.1 | 9.88 | <0.001 |
| [25]/[0] | 2.84 | 2.27-3.55 | 2.91 | <0.001 |
CI: Confidence Interval; PTC: Papillary Thyroid Cancer; *: Concentration of 5-Aza
Table 3: Rap1Gap mRNA expression levels in thyroid tissues and cell lines.
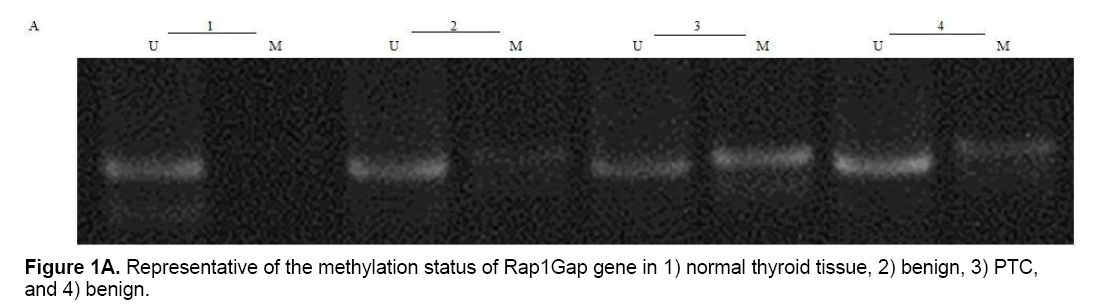
CpG24 hyper-methylation
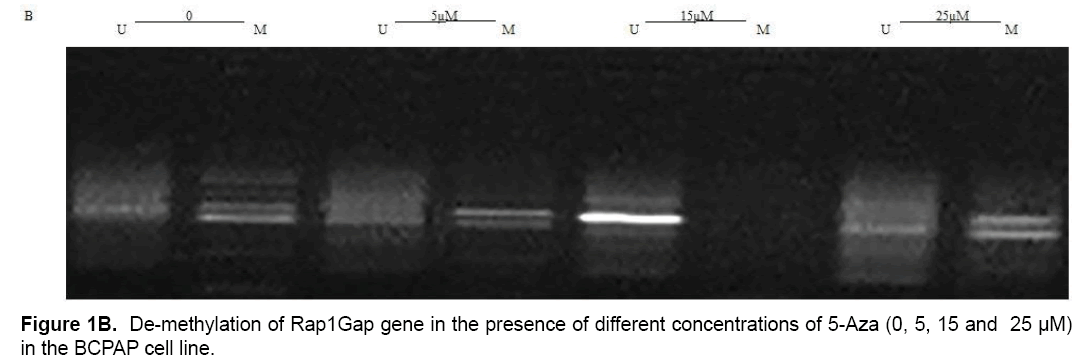
Present study showed that CpG24 is hyper-methylated in 82.3% of individuals with PTC, in 79.1% of benign nodules and only in 14% of normal thyroid tissues (p<0.001) (Table 4 and Figure 1A). According to these results, there was proposed that methylation status of CpG24 may be associated with PTC. Therefore, methylation pattern of this region could be considered as an epigenetic marker for papillary thyroid cancer, but this status is not necessarily associated with down-regulation of Rap1Gap gene. In this study, the levels of Rap1Gap gene expression were not correlated with methylation pattern of CpG24 in papillary thyroid cancer group (r=0.021; p=0.15). CpG24 was aberrantly methylated in B-CPAP and SW1736 cell lines. These cell lines were treated with 5-aza-2,-deoxycytidine in order to find, if the methylation pattern is associated with Rap1Gap gene expression and de-methylation of this region could induce the re-expression of this gene. Treatment with various concentrations of 5-Aza (5, 15, 25 μM) showed a dose dependent effect for 5-aza in both cell lines with optimum concentration of 15 μM (Table 3 and Figure 1B).
| UU | UM | MM | |
|---|---|---|---|
| Control (%) | 43 (86) | 7 (14) | 0 |
| Benign (%) | 10 (20.8) | 37 (77) | 1 (2.1) |
| PTC (%) | 6 (17.6) | 27 (79.4) | 1 (2.9)* |
UU: Un-Methylated; UM: Hetero-Methylated; MM: Homo-Methylated; PTC: Papillary Thyroid Cancer; p-value<0.001
Table 4: CpG24 methylation frequencies.
Discussion
The results of this study showed that Rap1Gap gene expression was not statistically different between healthy thyroid gland, benign nodule, and papillary thyroid cancer groups. In addition, hyper-methylation status of CpG24 was quantified; to the authors, knowledge, this is the first report investigating the CpG24 methylation in Iranian patients with papillary thyroid cancer. Accordingly, in PTC and benign groups, this region was 68.3% and 65.1% hyper-methylated more than the control group. CpG24 hyper-methylation could be considered as an epigenetic biomarker in PTC; however, it could not be necessarily considered as a reason of decreased Rap1Gap gene expression. Moreover, study on the BCPAP and SW1736 thyroid cancer cell lines indicated 5Aza-2deoxi cytidine as a de-methylating reagent that is able to reduce the aberrant hyper-methylation of CpG24 and re-express the Rap1Gap gene.
There are a few researches done about the role of Rap1Gap gene in determining thyroid cancer prognosis [14]. In this study we found that hyper- methylation of CpG24 is correlated with tumor size and lymph node involvement. Therefore, this epigenetic variation can be used as a prognosis biomarker in PTC patients, but more comprehensive studies are needed.
Actually, Rap1 as a member of the RAS superfamily interferes in the regulation of mitogenic and oncogenic pathways in thyroid tissues [15,16]. Previous studies have demonstrated that Rap1Gap is a tumor suppressor gene and its downregulation is associated with tumor malignancies [11,17,18]. The Rap1Gap gene expression has been reported to be an important marker for risk assessment of various cancers including renal cell carcinoma [19], leukemia [20], pancreatic cancer [17] and melanoma [21]. Several studies, which examined the association of Rap1Gap gene and cancer, give an insight into genetic variations leading to silencing of this gene; however, epigenetic patterns remain elusive. Aberrant DNA methylation is an important epigenetic alteration that is responsible for enhanced expression of proto-oncogenes as well as silencing of tumor suppressor genes leading to abnormal proliferation and de-differentiation of tumor cells [22].
Down-regulation of Rap1Gap was identified in melanoma cells by Zheng et al. [11]. They demonstrated a reduction of Rap1Gap expression and activity in cutaneous and metastatic melanoma tumors compared with human epidermal melanocytes. Their study also showed a re-expression in the Rap1Gap gene after treatment of melanoma cells with 5-Aza [11]. In another study, it was reported that the major mechanisms of Rap1Gap inactivation in thyroid cells are LOH and hyper-methylation in regulatory region [12]. Qiu et al. [20] have investigated the role of Rap1Gap gene in the leukemia cell differentiation, apoptosis and invasion. Their study indicated that Rap1Gap is able to promote differentiation, apoptosis and invasion of leukemia cells. Zhang et al. [23] identified Rap1Gap as a critical regulator of the aggressive tumor cells. They suggested that the Rap1Gap expression can influence on the migratory mechanisms of the operative in tumor cells [23]. Previous studies showed that CpG islands in the regulatory region of Rap1Gap gene are mostly methylated in cancer cells, but this aberrant DNA methylation is usually reversible [13,19,24]. Therefore, Rap1Gap gene could be considered as a potential target for cancer treatment.
Based on the above mentioned, our study strength includes its novelty for Iranian population. Despite to this strength, there were some limitations; first, methylation-specific PCR (MSP) was limited to provide the information at the level of individual CpG site, because there were various CpG islands in the regulatory region of the candidate gene. Second, it was impossible for us to find more thyroid cancer cell lines.
Conclusion
According to this study, we are able to conclude that aberrant DNA methylation in the CpG24 region, independent of Rap1Gap gene expression, could be considered as an epigenetic biomarker for papillary thyroid cancer.
Acknowledgement
We gratefully acknowledge Mr. Molaie for critical editing of English grammar and syntax of the manuscript and Miss. Fatemeh Eman for collecting samples. This work was funded by a grant from the Joundishapour University of Medical Sciences, Ahvaz, Iran (code: CMRC122).
References
- Zhao Z, Herman JG, Brock MV, et al. (2014). Methylation of DACT2 promotes papillary thyroid cancer metastasis by activating Wnt signaling. PLoS One. 9: e112336.
- Romei C, Elisei R. (2012). RET/PTC Translocations and clinico-pathological features in human papillary thyroid carcinoma. Front Endocrinol. 3: 54.
- Nikiforov Y E, Nikiforova M N. (2011). Molecular genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol. 7: 569-580.
- Nikiforov Y, Yip L, Nikiforova M. (2013). New strategies in diagnosing cancer in thyroid nodules: Impact of molecular markers. Clin Cancer Res. 19: 2283-2288.
- Yoo CB, Jones PA. (2006). Epigenetic therapy of cancer: Past, present and future. Nat Rev Drug Discov. 5: 37-50.
- Baylin SB, Jones PA. (2011). A decade of exploring the cancer epigenome - Biological and translational implications. Nat Rev Cancer. 11: 726-734.
- Dawson MA, Kouzarides T. (2012). Cancer epigenetics: From mechanism to therapy. Cell. 150: 12-27.
- Wang D, Zhang P, Gao K, et al. (2014). PLK1 and beta-TrCP-dependent ubiquitination and degradation of Rap1GAP controls cell proliferation. PLoS One. 9: e110296.
- Zhang Z, Mitra RS, Henson BS, et al. (2006). Rap1GAP inhibits tumor growth in oropharyngeal squamous cell carcinoma. Am J Pathol. 168: 585-596.
- Jo YS, Li S, Song JH, et al. (2006). Influence of the BRAF V600E mutation on expression of vascular endothelial growth factor in papillary thyroid cancer. J Clin Endocrinol Metab. 91: 3667-3670.
- Zheng H, Gao L, Feng Y, et al. (2009). Down-regulation of Rap1GAP via promoter hypermethylation promotes melanoma cell proliferation, survival and migration. Cancer Res. 69: 449-457.
- Zuo H, Gandhi M, Edreira MM, et al. (2010). Downregulation of Rap1GAP through epigenetic silencing and loss of heterozygosity promotes invasion and progression of thyroid tumors. Cancer Res. 70: 1389-1397.
- Banerjee R, Mani RS, Russo N, et al. (2011). The tumor suppressor gene rap1GAP is silenced by miR-101-mediated EZH2 overexpression in invasive squamous cell carcinoma. Oncogene. 30: 4339-4349.
- Li JJ, Zheng PCJ, Wang YZ. (2017). The correlation between DNA methylation and polymorphisms in the promoter region of the human telomerase reverse transcriptase (hTERT) gene with postoperative recurrence in patients with thyroid carcinoma (TC). World J Surg Oncol. 15: 114-117.
- De Falco V, Castellone M D, De Vita G, et al. (2007). RET/papillary thyroid carcinoma oncogenic signaling through the Rap1 small GTPase. Cancer Res. 67: 381-390.
- Wang Z, Dillon TJ, Pokala V, et al. (2006). Rap1-mediated activation of extracellular signal-regulated kinases by cyclic AMP is dependent on the mode of Rap1 activation. Mol Cell Biol. 26: 2130-2145.
- Zhang L, Chenwei L, Mahmood R, et al. (2006). Identification of a putative tumor suppressor gene Rap1GAP in pancreatic cancer. Cancer Res. 66: 898-906.
- Tsygankova OM, Prendergast GV, Puttaswamy K, et al. (2007). Downregulation of Rap1GAP contributes to Ras transformation. Mol Cell Biol. 27: 6647-6658.
- Kim WJ, Gersey Z, Daaka Y. (2012). Rap1GAP regulates renal cell carcinoma invasion. Cancer Lett. 320: 65-71.
- Qiu T, Qi X, Cen J, et al. (2012). Rap1GAP alters leukemia cell differentiation, apoptosis and invasion in vitro. Oncol Rep. 28: 622-628.
- Weiss J, Biwer B, Schliz M, et al. (1997). Clinicopathological and prognostic relevance of Rap1-GAP expression in melanocytic tumors. Arch Dermatol Res. 289: 573-577.
- Faam B, Ghaffari M A, Ghadiri A, et al. (2015). Epigenetic modifications in human thyroid cancer. Biomed Rep. 3: 3-8.
- Zhang X, Li D, Li M, et al. (2013). microRNA-146a targets PRKCE to modulate papillary thyroid tumor development. Int J Cancer. 134: 257-267.
- Dong X, Korch C, Meinkoth JL. (2011). Histone deacetylase inhibitors upregulate Rap1GAP and inhibit Rap activity in thyroid tumor cells. Endocr Relat Cancer. 18: 301-310.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences