The Green Fluorescent Protein (GFP) is a Vital Visual Marker in Citrus Transgene Research
Qifa Zheng, Chunxian Chen, Shu Huang, Xu Xiang, Young A. Choi & Fred G. Gmitter Jr
1Agriculture and Agri-Food Canada,Kentville Research Center,32 main street,Kentville,Nova Scotia,B4N 1J5,Canada
2Citrus Research and Education Center,University of Florida,700 Experiment Station Rd.,Lake Alfred,FL 33850 USA
3Institute of Fruit Tree Research,Guangdong Academy of Agricultural Science,Wushan,Guangzhou,Guangdong,510640 China
Abstract
pGREEN is a flexible Ti binary vector for Agrobacterium mediated transformation system [1]. The advantage of this vector is that optional combinations of selection and reporter gene cassettes can be optimally assembled to meet different transformation requirements. gfp (green fluorescent protein) gene driven by 35S-CaMV promoter was used as a reporter gene to enable transformant detection in vivo and the REDExtract-N-Amp Plant PCR Kit was applied as a high throughput approach for screening large amount of regenerated transformed plants in this report. With the designated vector pGK assembled using pGreen0029 and 35S-GFP, different phenotypes of transformation chimerism were observed and average transformation rates 7.75% and 2.03 % were acquired in term of consistent PCR results between the selectable and the reporter gene for empty vector and inserted recombinants respectively in transformation events of citrus grapefruits.
Key words
Agrobacterium,transformation,citrus,GFP (green fluorescent protein),PCR
1. Introduction
Genetic transformation is the direct approach for gene function characterization for any important clone selected from constructed bacterial libraries in genomic projects. Citrus is one of the most recalcitrant woody species to transform in gene engineering. For gene function evaluation for a bunch of gene candidates from genomic libraries,high transformation efficiency is required to acquire large amount of regenerated transformed plants and the high throughput reliable techniques are critical for screening those transformed plants without destructivity. We have compared several widely used expression vectors in our citrus transformation and excellent progress has been made for obtaining high transformation efficiency while in many cases,transformed plants will be damaged during transgene detection procedure and transgene verification won’t be done until other plants are grown enough size for sampling.
The green fluorescent protein (GFP) converts blue light emitted from aequorin in jellyfish to green light. The use of GFP marker protein has been described in bacteria [2],yeast [3],worm [4],fly [5] and mammals [6]. In plants,GFP has also been widely used as a visual marker gene that functions as a reporter of gene expression and permits the recovery of transformed plants [7].
pGREENs are a new set of flexible binary vectors for crop transformation newly developed by Roger Hellens et al. [8] and they have been optimised for the improvement of plant transformation via Agrobacterium. In pGREEN,the T-DNA region is flanked by BglII sites,which are useful in post-transformation analysis. The polylinker is based on pBluescript,and all associated plasmids and cassettes have been modified,to remove restriction sites included in this polylinker. Further unique sites HpaI and StuI are located internal to the Left and Right border sequence respectively. These blunt sites are designed to facilitate the cloning of selection and marker cassettes,which are flanked by blunt EcoRV sites. The npt II gene of pACYC 177 conveys resistance to kanamycin,and was used to select for plasmid transformation. Site-directed mutagenesis was used to remove the sites represented in the multiple cloning site,and an NcoI site was incorporated 3' of the gene,to facilitate convenient cloning. The RepA gene acts in trans upon the pSa Ori sequence. The RepA is therefore resident on a seperate plasmid pSoup. This plasmid can be co-electroporated with pGreen vector,or Agrobacterium cells with the plasmid can be made competent,for independent electroporation.
2. Materials and methods
2.1 Plant material
Seeds of grapefruit (Citrus paradisi Macf. ‘Duncan’) were sterilized with 5% bleach for 5 min and rinsed four times with sterile water. Sterile seeds were cultivated in G medium,i.e. ½ MS,03 mg/L NAA,2% sucrose and 0.8% phytoagar (pH5.7) at 26ºC under darkness for 4 weeks. On the transformation day,shoots of etiolated seedlings were collected and cut into fragments in 1 cm long as initial transformation explants.
2.2 Plasmid construction
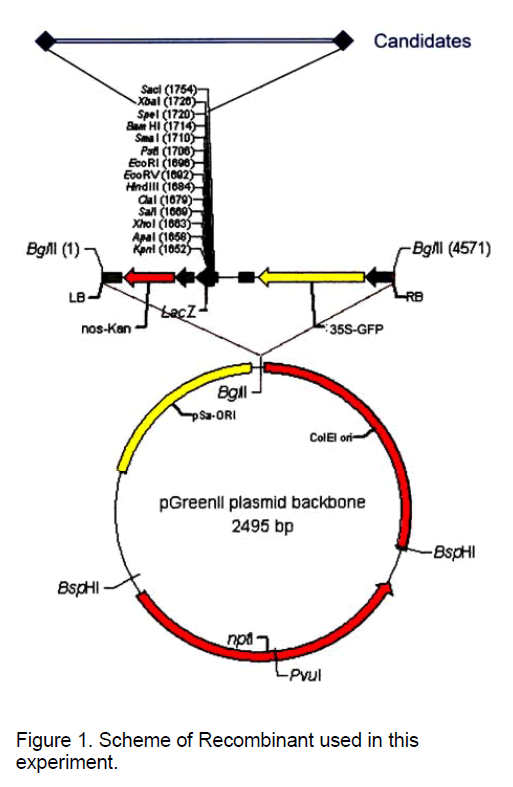
Recombinants containing genomic fragments of Poncirus were constructed by Dr. C.X. Chen by inserting selected DNA fragments into between Xba I and Spe I cloning site of the designed vector pGK,which contained the pGreenII backbone,the nos-Kan (nptII under the nos promoter for kanamycin resistance),the lacZ gene with the multiple cloning sites from pBlueScript,and GFP as the reporter,assembled with pGreen0029 and 35S-GFP (the GFP reporter gene driven by 35S-CaMV promoter) provided by Roger Hellens and Phil Mullineaux (Figure 1).
2.3 Agrobacterim transformation
1 μL target clone DNA (0.1-0.5 μg) was laid in TE buffer (pH 8.0) on top of 40 μL thawed frozen competent Agrobacterium AGL1 cells,on ice 1 min,DNA-competent cell mixture was pippetted in an ice cold 0.2 cm electroporation cuvette and electroporated in Bio-Rad Gene Pulser system. Parameters are 25 μF,400Ω,2.49 kv followed by a 8-9 ms delay. Immediately rinsed the cuvette with 0.8 mL LB antibiotic free medium and transferred the mixture to a sterile culture tube followed by incubating with shaking at 240 rpm,28C for 2.5 hrs. The electroporated cells were collected by centrifuge at 12,000 g for 2 min and the pellet was resuspended in 200 μL LB broth. Spread 25-50 μL of the culture on LB plate containing 50μg/mL of rifamycin,carbenicillin and kanamycin. The plates were incubated at 28°C for 2 days. Temperature should not be over 30°C which will result in loss of plasmid.
2.4 Plant transformation
Citrus transformation procedure is modified according to Yu et al. [9]. Streak Agrobacterium strain harboring recombinants or empty vector only on LB agar plates contained 50μg/mL of rifamycin,carbenicillin and kanamycin and incubate the above plates in incubator at 28°C for 2 days. Single colonies of A. tumefaciens was grown overnight at 28°C in 3 mL liquid LB medium (Invitrogen life technologies) supplemented with the same antibiotics to OD 1.0. Dilute bacterium to 1/100 vol/vol in 10 mL fresh LB broth with same antibiotics and 100 μM of the vir gene inducer acetosyringone (AS),and inoculate overnight to obtain culture of OD600 = 0.5-1.0. The pellet of A. tumefaciens was collected through centrifuge at 3000 g 10 minutes at room temperature. Re-suspend the bacterium pellet with 10 mL resuspended medium RM and centrifuge as above. Liquid bacterium resuspension medium (RM) is MS supplemented with 6-benzylaminopurine (6-BA) 3 mg/L,α-naphthaleneacetic acid (NAA) 0.1 mg/L and sucrose 50 g/L,pH=5.5. Recentrifuge and Resuspend the washed pellet with another 10 mL RM for transformation infection. Around 1 cm long cut epicotyl fragments from four weeks old etiolated seedlings were immersed in RM bacterium liquid for about 5 minutes,shaking slowly and frequently by forceps and then extra liquid was removed by autoclaved filter paper. The infected explants were placed horizontally on co-cultivation medium (CM) which is RM supplemented with 100 μM AS. Spare some non-infected and infected explants for setting up systematic controls. All co-cultivated plates were sealed with parafilm and placed at 26°C in the continuous darkness for 2.5 days. Explants were then transferred onto selection medium (SM) for selection and regeneration. Two steps were employed in selection phase,first in continue darkness for two weeks and then in the fluorescent light on the photoperiod of 16h/8h light/darkness for four weeks. Solid selection medium is MS supplemented with 6-BA 3 mg/L,NAA 0.4 mg/L,sucrose 50 g/L,cefotaxim (Cef) 450 mg /L,75 mg/L of kanamycin as a selection reagent and phytoagar 8g/L,pH=5.8. After four weeks of selection,agar was carefully removed from regenerated shoots in selection and regeneration medium by rinsing in tap water and they were grafted onto Carrizo rootstock grown in trays on chamber or in tubes for speeding up growth. MS basal salt Mixture (Sigma #M5519) was substituted for MS in this research.
2.5 GFP observation
Explants with small regenerated shoots grown on the parafilm-sealed culture plates containing selection medium were observed and images were captured under the digital fluorescence microscope. Shoots were then grafted onto rootstocks and marked accordingly for further growth and detection.
2.6 Identification of transgenic plants through PCR technique
All marked putative transformants were screened initially through PCR with the REDExtract-N-Amp Plant PCR Kits following manufacturer’s instruction (Sigma,Saint Louis,Missouri,USA). Briefly,a 0.5 cm leaf disc was punched from each plant using a standard one-hole paper punch and collected in a 2-ml collection tube followed by adding 100 μl of Extraction Solution. Close the tube and vortex briefly. Make sure the disk is covered by the Extraction Solution. Incubate samples at 95 ºC for 10 minutes. Add 100 μl of Dilution Solution and vortex to mix. Store the diluted leaf extract at 2-8 ºC until use. 4μl of the diluted extract was used for each PCR reaction.
Selectable marker gene npt II and reporter gene gfp were checked in all marked transformed plants. NptII gene-derived primers were 5’- TCGGCTATGA CTGGGCACAACAGA-3’ (24-mer) (forward),and 5’-AAGAAGGCGATAGAAGGCGATGCG-3’ (24-mer) (reverse),which are within nptII (conferring kanamycin resistance) coding region of npt II gene. PCR product is 722 bp. Primers derived from gfp coding region are available upon request.
Reaction parameters are at 94°C,3 min and then 94°C,2 min; 93°C,30 sec,55°C,90 sec,72°C,1.5 min,for 35 cycles; 72°C,7 min for final extension. 15 μL PCR products were loaded in 1.0 % agarose gel containing 0.1 μg/mL ethidium bromide for each sample and gel was run in 1×TAE at 7 v/cm. Images were captured with digital gel imaging system.
Figure 2 . GFP expression of transgenic shoots/tissue of grapefruit under stereomicroscope. 1: nontransgenic regenerated shoots on explants,clearly appeared red,negative control; 2-15: Transgenic shoots being verified by PCR,showing different phenotypes of expression of gfp gene,arrow points at callus containing GFP protein. Plants 7-12 showed sectorial expression of gfp in leaves from chimeric regenerated shoots. Plants 13,14 showed transgenic plants displaying bright green fluorescence. Plant 15 showed different types of transformants regenerated lined closely on the same cutting end of an explant.
3. Results and Discussion
3.1 Types of transgenic plants observed under the digital fluorescence microscope
GFP expression observation showed that there are some different phenotypes over the expression of gfp gene in regenerated shoots of transgenic grapefruit; some are chimeric (Figure 2 ). transformants appeared fully bright green fluorescence under the fluorescence stereomicroscope were selected from others for future functional testing of the target genes.
Figure 2. GFP expression of transgenic shoots/tissue of grapefruit under stereomicroscope. 1: nontransgenic regenerated shoots on explants, clearly appeared red, negative control; 2-15: Transgenic shoots being verified by PCR, showing different phenotypes of expression of gfp gene, arrow points at callus containing GFP protein. Plants 7-12 showed sectorial expression of gfp in leaves from chimeric regenerated shoots. Plants 13, 14 showed transgenic plants displaying bright green fluorescence. Plant 15 showed different types of transformants regenerated lined closely on the same cutting end of an explant.
3.2 Foreign gene identification in tranformants and estimation of transformation efficiency through REDExtract-N-Amp Plant PCR Kit
nptII gene and gus A gene were clearly detectable consistently in 66.67% (12 out of 18) of regenerated plants. nptII gene was detectable in another 16.67% (3 out of 18) of regenerated plants,while gus A gene was not. There were 16.67% of regenerated plants that were considered as escapes (Fig. 3).In term of consistent PCR analysis with both selectable marker and the reporter gene,transformation rate are approximately 7.75% and 2.03% for empty vector and recombinant containing genomic DNA inserts respectively (Table 1). Target genomic gene (s) can be detectable in only a portion of those plants that contained both selectable and reporter genes (data not shown). The REDExtract-N-Amp Plant PCR Kit enables us to look into each portion of the plant and up to one hundred transformants at a time in a day. More leaves discs from different leaves are recommended if the chimeric plants appear in the transformation events. The corresponding results depend on the type of the transgenic plants or on which portion of the chimeric leaves was sampled for PCR experiment. Expected target transgenic plants should be selected excluding the chimeric for accurate functional characterization of the candidate gene(s).
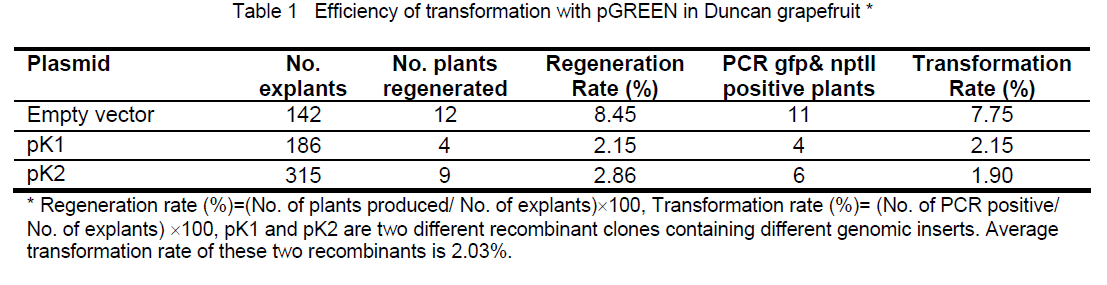
Figure 3. Electrophoresis images for PCR results amplified with a selectable gene npt II derived primers and a reporter gene gfp derived primers for transformed grapefruit plants. M: 1 kb DNA marker, the sizes of the DNA standards are indicated on left side of images. D: Duncan nontransformant, receptor plant background check, negative control P: plasmid pGREEN, PCR systematic check, positive control, H: DNA free water to check possible contamination of components for PCR 1-18: Regenerated plants showing consistent positive PCR results between selectable and reporter genes (espected transgenic plants, plants 1-9, 11-13) and inconsistent PCR results (nptII gene was detectable and gus A gene was undetectable in plant 10,15,16). Plants 16,17,18 were considered as escapes.
Acknowledgement
The authors would like to thank Ms Marjorie K. Wendell for her excellent technical assistance in this research. Thank Dr. Vladimir Orbovic,manager of Core Citrus Transformation Facility,at UF/IFAS CREC for his their helpful assistance in GFP observation.
References
- Hellens R.P.,Edwards E.A.,Leyland N.R.,et al. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation,Plant Mol. Bio.,42: 819-832.
- Lima A.O.,de S. (2001) Use of GFP (Green Fluorescent Protein) in a directed evolution approach of the beta-glucosidase A of Fervidobacterium sp. Piracicaba,SP. 2001. p115.
- Belhocine S. (2003) Establishing genetic modles in yeast for dissecting program cell death in eukaryotes. Chania CIHEAM IAMC. 2003. p118
- McMiller T.L.,Johnson C.M. (2005) Molecular characterization of HLH-17,a C-elegans bHLH protein required for normal larval development. Gene,356: 1-10.
- Yeh E.,Gustafson K.,Boulianne G.L. (1995) Green fluorescent protein as a vital marker and reporter of gene expression in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 92(15): 7036-3040.
- Iwasaki S.,Takada T.,Iida K,et al. (2000) GFP expression in mouse fetuses derived from aggregation of embryonic stem cells with diploid or tetraploid embryos. Journal of Mammalian Ova Research. 17(1): 15-22.
- Wahlroos T.,Susi P.,Tylkina L.,et al. (2003) Agrobacteriumumediated transformation and stable expression of the green fluorescent protein in Brassica rapa. Plant Physiology and Biochemistry,41(9): 773-778.
- Hellens R,Mullineaux P,Klee H. (2000) A guide to Agrobacterium binary Ti vectors,Trends in Plant Science,5(10): 446-451.
- Yu C.S.,Huang Z.,Deng C.,et al. (2002) Improving the efficiency of Agrobacterium-mediated transformation and plant regeneration in citrus. Plant Cell,Tissue,and Organ Culture,71: 147-155.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences