Study of Class 1 to 3 Integrons in Salmonella and Antimicrobial Resistance Pattern Isolated from Broiler
Mehrnoosh Doosti Irani, Mostafa Faghani* and Abbas Doosti
Mehrnoosh Doosti Irani1, Mostafa Faghani2* and Abbas Doosti1
1Biotechnology Research Center, Islamic Azad University, Shahre-Kord Branch, Shahre-Kord, Iran;
2Department of Animal Science, Faculty of Agriculture, Shahre-Kord Branch, Islamic Azad University, Shahre-Kord, Iran.
Received date: April 30, 2018; Accepted date: May 02, 2018; Published date: May 20, 2018
Citation: Irani MD, Faghani M, Doosti A. Study of Class 1 to 3 Integrons in Salmonella and Antimicrobial Resistance Pattern Isolated from Broiler Chicks. Electronic J Biol, 14:2
Abstract
High prevalent of multiple drug resistant (MDR) Salmonella is considered as a threat for human’s health. Integrons are one of the most important factors that can contribute to the occurrence of MDR bacteria. The aim of this study was to determine the prevalence of class 1, 2 and 3 integrons among Salmonella strains isolated from broiler chicks. This study was performed on 100 Salmonella isolated strains, collected from male and female broiler chicks samples in southwest of Iran. The prevalence of class 1-3 integrons were verified using specific primers by multiplex PCR assay. Also antimicrobial susceptibility testing by disk diffusion method was performed for each isolate. Screening of Salmonella isolates revealed the prevalence of class 1, 2 and 3 integrons (50%), (28%) and (48%), respectively. Based on the results of this study significant correlations were between MDR and integrons, this is a serious problem in human and veterinary medicine. According to these results Ampicillin was the most resistant antibiotics against Salmonella isolated strains. The resistance to Gentamicin and Tetracycline and Chloramphenicol has increased in the presence of integrons. The presence of all three classes of integrons and its direct connection with the MDR in Salmonella is concerned.
Keywords
Broiler chicks; Integrons; Iran; Multiplex PCR; Salmonella.
Introduction
Salmonella is a facultative intracellular pathogen that causes a variety of infectious diseases such as gastroenteritis. It is one of the serious health problems worldwide and causes global morbidity and mortality [1]. Human salmonellosis is one of the most common and economically important zoonotic diseases and can affect all species of domestic animals, especially pigs and poultry. High prevalent of multidrug resistance (MDR) Salmonella are considered as a threat for human’s health [2]. The genus of Salmonella belongs to the Enterobacteriaceae family and includes more than 2500 different serotypes or serovars [3]. These bacteria are rod-shaped, gram-negative, mesophilic, non-spore-forming, predominantly motile and peritrichous flagella [2]. Salmonella is an important cause of bacterial food borne disease in industrialized countries and it is responsible for significant economic losses and serious health problems [4]. Although most Salmonella infection is limited to uncomplicated gastroenteritis that seldom requires antimicrobial treatment and severity of gastroenteritis and instead may result in prolonged fecal excretion and emergence of resistant strains [5]. Antibiotic resistance of Salmonella is a major threat to human health and veterinary worldwide [6,7].Currently, the indiscriminate use and misuse of antibiotics has facilitated the emergence of resistance in many Salmonella serovars [8]. Virulence factors and resistance may be present on chromosomes, plasmids, transposons, integrons or phage located and they are able to integrate and express genes coding for antibiotic resistance [1].
Integrons are genetic elements that lead to various combinations of MDR pattern [9]. Horizontal transfers of integrons are an important factor of MDR in gramnegative bacteria [10,11]. Because of integrons are not capable of horizontal gene transfer themselves gene cassettes are necessary for combination in integron and often associated with transposons or plasmids. Over 80 gene cassettes and 8 classes of integrons have been characterized to date [12,13]. Class 1 and 2 integrons were found to be the most common and are referred to as resistance integrons but class 3 integrons are rare [14]. Classes 1 integrons are the most abundant type of integrons which are important for the creation and transfer of antibiotic resistance [1]. All integrons characterized are composed of three key elements necessary: a gene encoding an integrase belonging to the tyrosinerecombinase family (intI), a primary recombination site (attI) and promoter (Pc) [15]. The aim of this study was to prevalence of class 1, 2 and 3 integrons of Salmonella that isolated from broiler chicks and their relationship with MDR and also associations between integrons with drug resistances.
Materials and Methods
Bacterial isolation and identification
The number of 100 samples was collected randomly from male and female broiler chicks in southwest of Iran. After culturing and isolating bacteria, genomic DNA for the polymerase chain reaction (PCR) assays was performed by the boiling method [1].
PCR
Discussion
The presence of class 1-3 integrons in Salmonella isolates were tested by multiplex PCR using specific primers for integrases genes of the integron, intI1, intI2, and intI3 introduced by Kargar et al. [11]. Each 50 μL reaction mixtures contained 10x reaction buffer, 0.5 μL dNTP mixture and 1.5 μL MgCl2 along with 0.5 unit TaqDNA polymerase, 0.75 μL of each primer (class 1, 2 and 3 integrins) and 2.5 μL of template DNA. For negative control a tube containing PCR reaction without any DNA template was used [11].
To identify genes related to the genera and species of enteritidis in Salmonella SefA primers were used. Primer sequences and PCR conditions were shown in Table 1. PCR amplification was performed in a 25 μL reaction volume containing 1 μg of genomic DNA, 1 μM of each primer (forward and reverse of Sef A), 2 mM MgCl2, 200 μMdNTP, 2.5 μLof 10x PCR buffer and 1 unit of Taq DNA polymerase (CinnaGen Co, Iran). The PCR products were analyzed by electrophoresis through 1.5% agarose gel, after which the gel was stained with ethidium bromide and photographed [16].
| Primer name | Primer sequence | Observed size (bp) | PCR conditions |
|---|---|---|---|
| IntI 1-F | ACATGCGTGTAAATCATCGTC | 483 | 5 min at 94°C 32 cycles of 1 min at 94°C 1 min at 59°C 1 min at 72°C 10 min at 72°C |
| IntI 1-R | GGTCAAGGATCTGGATTTCG | ||
| IntI 2-F | CACGGATATGCGACAAAA AGG | 788 | |
| IntI 2-R | ACATGCGTGTAAATCATC GTC | ||
| IntI 3-F | TGTTCTTGTATCGGCAGGTG | 588 | |
| IntI 3-R | AGTGGGTGGCGAATGAGTG | ||
| Sef A-F | TGCTATTTTGCCCTGTACACTCG | 214 | 5 min at 94°C 32 cycles of 1 min at 94°C 1 min at 63°C 1 min at 72°C 10 min at 72°C |
| Sef A-R | TTCGGGGAGACTATACCTACAG |
Table 1: Primers and PCR conditions used in this study.
Antimicrobial susceptibility testing
Antibiotic resistance test were performed using standard Bauer-Kirby disk diffusion method for all 100 samples. Antimicrobial agents tested were Trimethoprim-sulfamethoxazole (SXT=1.25/23.75 μg), Gentamicin (GM=10 μg), Ciprofloxacin (CP=5 μg), Nalidixic acid (NA=30 μg), Chloramphenicol (C=30 μg), Tetracycline(tet=30 μg), Clindamycin (DA=2 μg), Ampicillin (AMP=10 μg), Cephalexin (CL=30 μg) (Padtanteb, Tehran, Iran). Diameters of the inhibition zones were interpreted according to the Clinical and Laboratory Standards Institute (CLSI).
Statistical analysis
All data were analyzed by using MS Excel 2010 and SPSS software (Version 20. SPSS Inc., USA). At first, the frequency of resistance for each antibiotic (include 3 group: sensitive, semi-sensitive and resistance) was calculate and then the significant difference between this 3 groups at each integron for antibiotics was calculated using Chi-square and Fisher’s exact tests.
Results
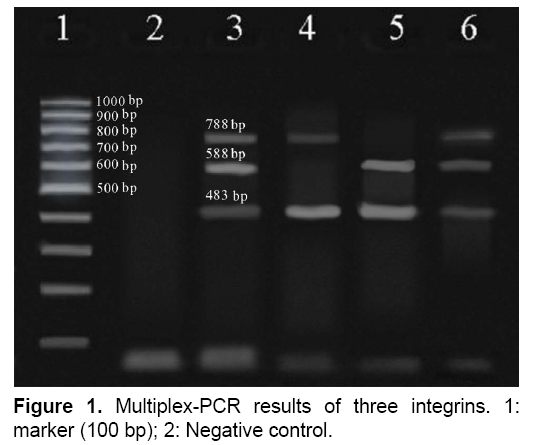
The result of multiplex-PCR results was showed in Figure 1, the size bands for integron class 1, 2 and 3 was 483 bp, 788 bp and 588 bp, respectively.
The PCR results in Figure 2, showed that bands 214 bp of the Sef A primers. Specific PCR analysis showed twenty-nine (29%) out of 100 isolates were species of S. enteritidis.
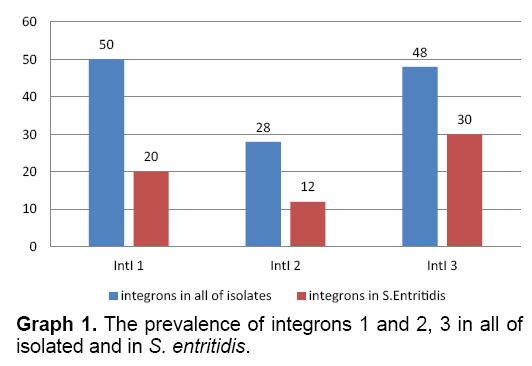
Based on the results of multiplex-PCR, the frequency of integrons 1, 2 and 3 in all of isolated samples were estimated as 50, 28 and 48 percent, respectively. The prevalence 3 groups integrin in S. enteritidis were 20, 12 and 30 percent, respectively (Graph 1).
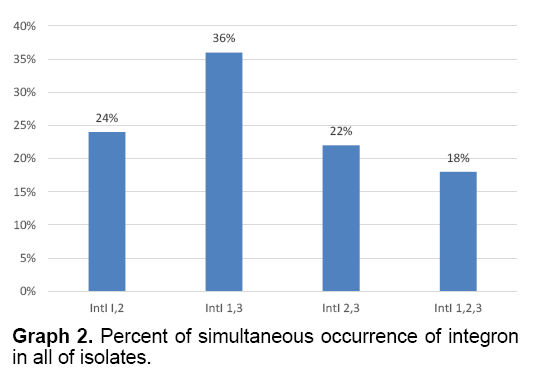
The present of simultaneous occurrence of integrons 1 and 2 were 24%, 1 and 3 were 36%, 2 and 3 were 22% and present of simultaneous occurrence of integrons 1, 2 and 3 were 18% (Graph 2).
The antibiotic resistance results indicated that all isolates as well as samples of positive integrons was sensitive to Ciprofloxacin and Trimethoprimsulfamethoxazole. In addition in all isolates 90% were resistant Clindamycin, 85% were resistant to Ampicillin, 57% resistant to Cephalexin, 5% resistant to Gentamicin and Tetracycline. However, the interpretation of the results show 100% of strains that have class 1, 2 and 3 integrons were resistant to Cephalexin, Clindamycin and Ampicillin.
According to Table 2, the antibiotic resistance to Gentamycin and Tetracycline in samples without IntI 1 were 2.6% and 2.5%, while examples of IntI1- positive have increased by 16% and the difference is significant (p<0.01). The strains intermediate resistant to Chloramphenicol in the samples that the IntI 1 were negative, were 3.8% while species intermediate resistant to Chloramphenicol were 72% in the samples IntI1-positive.
| Antimicrobial Agent | Antimicrobial resistance | R (%) | I (%) | S (%) | P value |
|---|---|---|---|---|---|
| Trimethoprim-sulfamethoxazole | IntI 1-positive | 0 | 0 | 100 | NS |
| IntI 1-negative | 0 | 0 | 100 | ||
| Ciprofloxacin | IntI 1-positive | 0 | 0 | 100 | NS |
| IntI 1-negative | 0 | 0 | 100 | ||
| Gentamicin | IntI 1-positive | 16 | 84 | 0 | ** |
| IntI 1-negative | 2.6 | 11.2 | 86.2 | ||
| Nalidixic acid | IntI 1-positive | 0 | 100 | 0 | NS |
| IntI 1-negative | 0 | 100 | 0 | ||
| Chloramphenicol | IntI 1-positive | 0 | 72 | 28 | ** |
| IntI 1-negative | 0 | 3.8 | 96.2 | ||
| Tetracycline | IntI 1-positive | 16 | 0 | 84 | * |
| IntI 1-negative | 2.5 | 0 | 97.5 | ||
| Clindamycin | IntI 1-positive | 100 | 0 | 0 | NS |
| IntI 1-negative | 100 | 0 | 0 | ||
| Ampicillin | IntI 1-positive | 100 | 0 | 0 | NS |
| IntI 1-negative | 100 | 0 | 0 | ||
| Cephalexin | IntI 1-positive | 100 | 0 | 0 | NS |
| IntI 1-negative | 100 | 0 | 0 |
R _ ResistantÃÆÃÅÃâââ¬Âº I _ Intermediate resistantÃÆÃÅÃâââ¬ÂºS _ susceptibleÃÆÃÅÃâââ¬ÂºNS _ Resistance difference was not significantÃÆÃÅÃâââ¬Âº** _ Resistance difference was significant (p<0.01)ÃÆÃÅÃâââ¬Âº* _ Resistance difference was significant (p<0.05)
Table 2: Comparison of class1integrons effect on the resistance to different antibiotics.
The resistance to Gentamicin were 32.4% from samples IntI 2-negative and this were 35.7% from strains positive IntI2. The resistance to Tetracycline was 21.4% in IntI2-positive while in IntI2-negative was 3.9% (Table 3).
| Antimicrobial Agent | Antimicrobial resistance | R (%) | I (%) | S (%) | P value |
|---|---|---|---|---|---|
| Trimethoprim-sulfamethoxazole | IntI 2-positive | 0 | 0 | 100 | NS |
| IntI 2-negative | 0 | 0 | 100 | ||
| Ciprofloxacin | IntI 2-positive | 0 | 0 | 100 | NS |
| IntI 2-negative | 0 | 0 | 100 | ||
| Gentamicin | IntI 2-positive | 35.7 | 64.3 | 0 | ** |
| IntI 2-negative | 32.4 | 0 | 67.6 | ||
| Nalidixic acid | IntI 2-positive | 0 | 100 | 0 | NS |
| IntI 2-negative | 0 | 100 | 0 | ||
| Chloramphenicol | IntI 2-positive | 0 | 42.9 | 57.1 | NS |
| IntI 2-negative | 0 | 26.5 | 73.5 | ||
| Tetracycline | IntI 2-positive | 21.4 | 0 | 78.6 | ** |
| IntI 2-negative | 3.9 | 0 | 96.1 | ||
| Clindamycin | IntI 2-positive | 100 | 0 | 0 | NS |
| IntI 2-negative | 100 | 0 | 0 | ||
| Ampicillin | IntI 2-positive | 100 | 0 | 0 | NS |
| IntI 2-negative | 100 | 0 | 0 | ||
| Cephalexin | IntI 2-positive | 100 | 0 | 0 | NS |
| IntI 2-negative | 100 | 0 | 0 |
R _ ResistantÃÆÃÅÃâââ¬Âº I _ Intermediate resistantÃÆÃÅÃâââ¬Âº S _ susceptibleÃÆÃÅÃâââ¬ÂºNS _ Resistance difference was not significantÃÆÃÅÃâââ¬Âº** _ Resistance difference was significant (p< 0.01)ÃÆÃÅÃâââ¬Âº* _ Resistance difference was significant (p<0.05)
Table 3: Comparison of class 2 integrons effect on the resistance to different antibiotics.
Interestingly, 21.3% from strain that have IntI3 shown antibiotic resistance to Gentamicin while none have shown the strains without integrons 3 resistance to these antibiotics. In addition resistance to Tetracycline has increased from 2.4% in strains IntI3-negative to 17% in IntI3-positive (Table 4).
| Antimicrobial Agent | Antimicrobial resistance | R (%) | I (%) | S (%) | P value |
|---|---|---|---|---|---|
| Trimethoprim-sulfamethoxazole | IntI 3-positive | 0 | 0 | 100 | NS |
| IntI 3-negative | 0 | 0 | 100 | ||
| Ciprofloxacin | IntI 3-positive | 0 | 0 | 100 | NS |
| IntI 3-negative | 0 | 0 | 100 | ||
| Gentamicin | IntI 3-positive | 21.3 | 78.7 | 0 | ** |
| IntI 3-negative | 0 | 16.9 | 83.1 | ||
| Nalidixic acid | IntI 3-positive | 0 | 100 | 0 | NS |
| IntI 3-negative | 0 | 100 | 0 | ||
| Chloramphenicol | IntI 3-positive | 0 | 57.4 | 42.6 | ** |
| IntI 3-negative | 0 | 14.5 | 85.5 | ||
| Tetracycline | IntI 3-positive | 17 | 0 | 83 | * |
| IntI 3-negative | 2.4 | 0 | 97.6 | ||
| Clindamycin | IntI 3-positive | 100 | 0 | 0 | NS |
| IntI 3-negative | 100 | 0 | 0 | ||
| Ampicillin | IntI 3-positive | 100 | 0 | 0 | NS |
| IntI 3-negative | 100 | 0 | 0 | ||
| Cephalexin | IntI 3-positive | 100 | 0 | 0 | NS |
| IntI 3-negative | 100 | 0 | 0 |
R _ ResistantÃÆÃÅÃâââ¬Âº I _ Intermediate resistantÃÆÃÅÃâââ¬Âº S _ susceptibleÃÆÃÅÃâââ¬ÂºNS _ Resistance difference was not significantÃÆÃÅÃâââ¬Âº** _ Resistance difference was significant (p< 0.01)ÃÆÃÅÃâââ¬Âº* _ Resistance difference was significant (p<0.05)
Table 4: Comparison of class3 integrons effect on the resistance to different antibiotics.
Discussion
Multidrug-resistant bacteria pathogens such as Salmonella have become a major human health concern in developed and developing countries [17- 19]. Salmonella has a broad vertebrate host range exists and most Salmonella disease are zoonotic [6,20]. The pollution of food products and the infection of foods of animal origin with Salmonella isolates carrying antibiotic resistance genes is an increasing concern [21]. Salmonella transmit by commercial industrial production of eggs, meat and food chains and can transfer resistance genes to human internal flora [1,6,20]. In recent years MDR among Salmonella is increasing, there for monitoring genotypic and phenotypic resistance to antibiotics in Salmonella is important for the public health [22,23]. In Iran, Japan and China have reported a high frequency of resistance to nalidixic acid and ciprofloxacin for S. enterica and S. infantis [24-27]. Asgharpour et al. [1] founded that all Salmonella isolates with MDR patterns had class 1integron gene and showed that all of them were sensitive to Nalidixic acid, Streptomycin and Tetracycline [1]. In similar study, Kargar et al. [11] prevalent of class 1, 2 and 3 integrons in Escherichia coli were estimated as 78.26%, 76.81% and 26.09% [11]. The study in Armenia had shown the high presence of class I integrons and associated drug resistance among the S. enterica isolates [19]. According to other studies from Iran, the occurrence of infection and MDR to Salmonella is increasing in human and class 1 integrons more prevalent than class 2 in Salmonella isolates and to be associated with MDR [18,28]. The study in 2008, shown the high frequency of occurrence class 1 integrons in Salmonella strains and a strong association of with identified resistance antibiotics were demonstrated [29].
In this study similar to other reports, the prevalence of intI1 (50%) was higher than the other two classes, in contrast to other studies, intI3 (48%) is the second highest rate and intI2 (28%) had the lowest prevalence among Salmonella isolated. The increasing presence intI3 in Salmonella is concerned. Presence of integrons in S. enteritidis is more than total samplers. In this study, intI2 associated with an integrons or two others. Based on the results of the antibiogram test, resistance to antibiotics direct associated with presence of integrons. Resistance to antibiotics Gentamicin, Tetracycline and Chloramphenicol associated with the presence of integrons. Increasing the presence of integrons in Salmonella and its relation to resistance to these antibiotics is concerned. Attention to the resistance of all samples of Salmonella isolates to antibiotics Clindamycin, Clindamycin and Cephalexin the use of antibiotics is not recommended in the treatment of Salmonella.
Conclusion
In conclusions, receipt and dissemination of antibiotic resistance genes in integrons has potential role in distribution of MDR. The results showed high prevalence class 1, 2 and 3 integrons and also high percentage of antibiotic resistance in Salmonella.
Authors’ Contributions
AD conceived and designed the study. MDI carried out the laboratory work. MF participated in statistical analysis and procedures. NZJ coordinated and participated in designing the study. All the authors read and approved the final version.
Acknowledgement
The authors give special thanks and express their deep sense of gratitude to the staff of the Biotechnology Research Center of Islamic Azad University of Shahre-Kord Branch for their effort.
References
- Asgharpour F, Rajabnia R, Shahandashti EF, et al. (2014). Investigation of class I integron in Salmonella infantis and its association with drug resistance. Jundishapur J Microbiol. 7: e10019.
- Pal J, Raj R, Kumar J, et al. (2014). Food borne illness outbreaks associated with consumption of seafood. Life Sci Lealf. 51: 4-9.
- Chu C, Feng Y, Chien AC, et al. (2008). Evolution of genes on the Salmonella virulence plasmid phylogeny revealed from sequencing of the virulence plasmids of S. enterica serotype Dublin and comparative analysis. Genomics. 92: 339-343.
- Capalonga R, Ramos RC, Both JMC, et al. (2014). Salmonella serotypes, resistance patterns and food vehicles of salmonellosis in southern Brazil between 2007 and 2012. J Infect Dev Ctries. 8: 811-817.
- Ranjbar R, Salimkhani E, Sadeghifard N, et al. (2007). An outbreak of gastroenteritis of unknown origin in Tehran, July 2003. Pak J Biol Sci. 10: 1138-1140.
- Feasey NA, Dougan G, Kingsley RA, et al. (2012). Invasive non-typhoidal Salmonella disease: An emerging and neglected tropical disease in Africa. Lancet. 379: 2489-2499.
- Forshell LP, Wierup M. (2006). Salmonella contamination: A significant challenge to the global marketing of animal food products. Rev Sci Tech Off Int Epiz. 25: 541-554.
- Angulo FJ, Mølbak K. (2005). Human health consequences of antimicrobial drug-resistant Salmonella and other foodborne pathogens. Clin Infect Dis. 41: 1613-1620.
- Povilonis J, ŠeputienÃÆââ¬Å¾Ãâââ¬â V, RuÃÆââ¬Â¦Ãâþauskas M, et al. (2010). Transferable class 1 and 2 integrons in Escherichia coli and Salmonella enterica isolates of human and animal origin in Lithuania. Foodborne Pathog Dis. 7: 1185-1192.
- Colli, CM, Kim MJ, Partridge SR, H. et al. (2002). Characterization of the class 3 integron and the site-specific recombination system it determines. J Bacteriol. 184: 3017-3026.
- Kargar M, Mohammadalipour Z, Doosti A, et al. (2014). High prevalence of class 1 to 3 integrons among multidrug-resistant diarrheagenic Escherichia coli in Southwest of Iran. Osong Public Health Res Perspect. 5: 193-198.
- Gillings M, Boucher Y, Labbate M, et al. (2008). The evolution of class 1 integrons and the rise of antibiotic resistance. J Bacteriol. 190: 5095-5100.
- Xu H, Davies J, Miao V. (2007). Molecular characterization of class 3 integrons from Delftia spp. J Bacteriol. 189: 6276-6283.
- Jin Y, Ling J. (2009). Prevalence of integrons in antibiotic-resistant Salmonella spp. in Hong Kong. Jpn J Infect Dis. 62: 432-439.
- Mazel D. (2006). Integrons: Agents of bacterial evolution. Nat Rev Microbiol. 4: 608-620.
- Doosti, A, Dehkordi PG, Rahimi E. (2014). Molecular assay to fraud identification of meat products. J Food Sci Technol. 51: 148-152.
- Chen S, Zhao S, White DG, et al. (2004). Characterization of multiple-antimicrobial-resistant Salmonellaserovars isolated from retail meats. Appl Environ Microbiol. 70: 1-7.
- Naghoni A, Ranjbar R, Tabaraie B, et al. (2010). High prevalence of integron-mediated resistance in clinical isolates of Salmonella enterica. Jpn J Infect Dis. 63: 417-421.
- Zakharyan MA, Sedrakyan K, Arakelova A, et al. (2014). Antibiotic resistance and occurrence of class 1 integrons in clinical isolates of Salmonella enterica. Glob J Immuno Allerg. 1: 44-48.
- Anjum MF, Choudhary S, Morrison V, et al. (2011). Identifying antimicrobial resistance genes of human clinical relevance within Salmonellaisolated from food animals in Great Britain. J Antimicrob Chemother. 66: 550-559.
- Lopes GV, Michael GB, Cardoso M, et al. (2014). Identification and characterization of Salmonella enterica subsp. enterica serovar Derby isolates carrying a new aadA26 gene cassette in a class 1 integron obtained at pig slaughterhouses. FEMS Microbiol Lett. 356: 71-78.
- Hoelzer KA,. Switt IM, Wiedmann M. (2011). Animal contact as a source of human non-typhoidal salmonellosis. Vet Res. 42: 1.
- Ribeiro VB, Lincopan N, Landgraf M, et al. (2011). Characterization of class 1 integrons and antibiotic resistance genes in multidrug-resistant Salmonella enterica isolates from foodstuff and related sources. Braz J Microbiol 42: 685-692.
- Dahshan H T, Chuma F, Shahada M, et al. (2010). Characterization of antibiotic resistance and the emergence of AmpC-producing Salmonella infantis from pigs. J Vet Med Sci. 72: 1437-1442.
- Morshed R, Peighambari SM. (2010). Drug resistance, plasmid profile and random amplified polymorphic DNA analysis of Iranian isolates of Salmonella enteritidis. New Microbiol. 33: 47.
- Rahmani MSM, Peighambari CA, Svendsen LM, et al. (2013). Molecular clonality and antimicrobial resistance in Salmonellaenterica serovars Enteritidis and Infantis from broilers in three Northern regions of Iran. BMC Vet Res. 9: 1.
- Yan HL, Li MJ, Alam S, et al. (2010). Prevalence and antimicrobial resistance of Salmonellain retail foods in northern China. Int J Food Microbiol. 143: 230-234.
- Bozorgmehri F, Hassanzadeh MM, Emaddi Chashni S, et al. (2016). Characterization of the Salmonella isolates from backyard chickens in north of Iran, by serotyping, multiplex PCR and antibiotic resistance analysis. Arch Razi Inst. 64: 77-83.
- Rao S, Maddox CW, Hoien-Dalen P, et al. (2008). Diagnostic accuracy of class 1 integron PCR method in detection of antibiotic resistance in Salmonellaisolates from swine production systems. J Clin Microbiol. 46: 916-920.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences