Role of CBF/DRFBs in ABA Signalling during Cold Stress in Plants
Gunjan Garg, Durvesh Kumar
Gunjan Garg* and Durvesh Kumar
School of Biotechnology, Gautam Buddha University, Greater Noida, India.
Received date June 10, 2016; Accepted date August 08, 2016; Published date August 15, 2016
Citation: Garg G, Kumar D. Role of CBF/DRFBs in ABA signalling during cold stress in plants. Electronic J Biol, S1.
Abstract
Abiotic stresses, such as oxidative stress are serious threats to agriculture and the natural status of the environment. Recently, research into the molecular mechanism of stress responses especially on genetic modification in plants for stress tolerance has been started and it showing promising results once applies to agriculturally and ecologically important plants. This review examines the different classes of proteins involved in ABA signalling pathway during oxidative stress. The phytohormones ABA is produced by plants under cold and drought stress conditions and plays an important role in mediating stress tolerance. Because cold stress is related to ABA responses, therefore more than 100 genes are up regulated by cold stress, out of which few genes are necessarily need to be associated with cold tolerance. Many cold stress induced genes are activated by transcriptional activators called C-repeat binding factors (CBF; also known as DREB). In response to the cold, multiple regulatory pathways are also activated in plants. They involved several other transcription factors like: ICE1 (Inducer of CBF Expression 1). ICE1-CBF pathway is critical for the regulation of the cold-responsive transcriptome and acquired freezing tolerance.
Keywords
ABA; Cold stress; Oxidative stress; CBF (C-repeat binding factors).
1. Introduction
Stress is defined as mechanical pressure per unit area applied to an object. In response to the applied stress, an object undergoes a chance in the dimension, which is also known as strain. As plants are sessile, it is tough to measure the exact force exerted by stresses and therefore in biological terms it could be considered as ‘stressor’ (Figure 1). Plants generate stress specific responses for surviving in the stress condition. Tolerance of plants to oxidative stress such as cold, drought and salinity is triggered by different classes of proteins which are involve in signalling pathways in plants. Identification of different classes of proteins involved in ABA (Abscisic acid) signalling pathway during oxidative stress, which are helpful for production of new and improved stress tolerant plants. Abscisic acid (ABA) is an important phytohormone for plant growth and development. It plays an important role in integrating various stress signals and mimics the effect of a stress condition on plants. Main function of ABA seems to be the regulated the water balance and osmotic stress tolerance in plants. ABA level rise up under stress condition, which is mainly due to the induction of genes for enzymes responsible for ABA biosynthesis. The abiotic stress-induced activation of many ABA biosynthetic genes such as: Zeaxanthin Oxidase (ZEP), 9-cis-Epoxycarotenoid Dioxygenase (NCED), ABA-Aldehyde Oxidase (AAO) and Molybdenum Cofactor Sulphurase (MCSU). All the genes are regulated through calcium-dependent phosphorylation pathway. Some species of plant (e.g. Arabidopsis,) showed rapid expression of the CBF transcriptional activator (also known as DREB1 proteins) in response to low temperature followed by induction of the CBF regulon (CBF- target genes), which contribute to an increase in freezing tolerance (Thomashow). Genes that encode CBF/DREBs, are stress genes, because they are induced by stress condition and ABA. These genes were show various area including COR (cold regulated), LTI (lowtemperature induced), KIN (cold induced) and RD (response to dehydration). When a plant goes to low temperature, the transcriptome levels are increase, increased the expression of proteins in plants. When plants go to high temperature transcriptome levels are decrease shown non acclimated condition, decreased the expression of proteins.
2. Abiotic Stress and Its Signal Transduction Pathways
Abiotic stresses, such as drought, salinity, extreme temperatures, chemical toxicity and oxidative stress are serious threats to agriculture and the natural status of the environment.
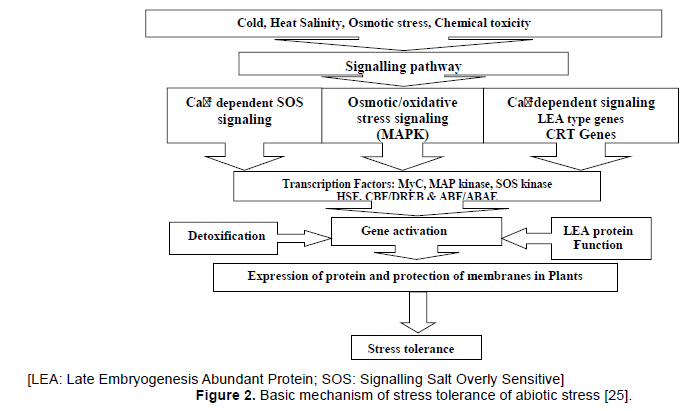
Plants have developed several biochemical, physiological and metabolite strategies in order to combat such as abiotic stresses. The signal transduction pathways in plants under environmental stress have been divided into major categories: (1) Osmotic/ oxidative stress signaling that makes use of mitogen activated proteins in kinase (MAPK) modules; (2) Ca+² dependent signaling that leads to activation of LEAtype genes such as Dehydration Responsive Elements (DRE)/Cold Responsive Sensitive Transcription Factors (CRT) class of genes (Figure 2).
2.1 Gene signalling under stress condition:
Some genes which are responsible for signalling in stress condition in plants such as MyC, MAP kinase and SOS kinase, Phospholipases, Transcriptional factors such as HSF, and the CBF/DREB and ABF/ ABAE families are involved in signalling cascades and in transcriptional control [1-3]. DREBs belong to induced by cold and dehydration, respectively. The DREBs are apparently involved in biotic stress signalling pathway. It has been possible to engineer stress tolerance in transgenic plants by manipulating the expression of DREBs. DREB/CBF proteins are encoded by AP2/EREBP multigene families and mediate the transcription of several genes such as rd29A, rd17, cor6.6, cor15a, erd10,kin2 and others in response to cold and water stress [4-7]. Jaglo- Ottosen et al. showed that increased expression of Arabidopsis CBF1 induces the expression of the cold-regulated genes cor6.6, cor15a, cor47 and cor78, and increased the freezing tolerance of nonacclimated Arabidopsis plants (Tables 1 and 2) [8].
3. Cold-Stress-Responsive Transcriptome and Freezing Tolerance Regulatory Pathway
| Genes | Species | References |
|---|---|---|
| CBF1 | Arabidopsis thaliana | Jaglo-Ottosen et al.[8]. |
| DREB1A | Arabidopsis thaliana | Kasuga et al. [13]. |
| CBF3 | Arabidopsis thaliana | Kasuga et al. [13]. |
| CBFs | Brassica napus | Jaglo et al. [8] |
| CBF1 | Lycopersiconesculentum | Hsieh et al. [14]. |
| CBF4 | Arabidopsis thaliana | Haake et al.[12]. |
| AtMYC2 andAtMYB2 | Arabidopsis thaliana | Abe et al. [15]. |
Table 1.List of genes responsible for stress signalling in plants.
| DREB TFS | Species | Accession no. | Stress responses | References |
|---|---|---|---|---|
| DREB1A | Arabidopsis thaliana | AB007787 | Cold | Lee et al. [14]. |
| DREB2A | Arabidopsis thaliana | AB007790 | Drought, salt, ABA | Lee et al. [16] |
| DREB2C | Arabidopsis thaliana | At2g40340 | Cold | Lee et al. [14] |
| CBF1 | Arabidopsis thaliana | U77378 | Cold | Gilmouret al. [8] |
| CBF2 | Arabidopsis thaliana | AF074601 | Cold | Gilmour et al. [8] |
| CBF3 | Arabidopsis thaliana | AF074602 | Cold | Gilmour, et al. [8] |
| CBF4 | Arabidopsis thaliana | AB015478 | Drought, ABA | Haake et al. [12] |
| OsDREB1A | Oryzasativa | AF300970 | Cold, Salt | Dubouzet et al. [18]. |
| OsDREB1B | Oryzasativa | AF300972 | Cold | Dubouzet et al. [18]. |
| OsDREB1C | Oryzasativa | AP001168 | Drought, Salt, cold, ABA | Dubouzet et al. [18]. |
| OsDREB1D | Oryzasativa | AB023482 | None | Dubouzet et al. [18]. |
| OsDREB2A | Oryzasativa | AF300971 | Drought, salt, cold, ABA | Dubouzet et al. [18]. |
| OsDREB1F | Oryzasativa | Drought, salt, Cold, ABA | Wang et al. [17]. | |
| OsDREB2B | Oryzasativa | Heat, cold | Matskura et al. [19]. | |
| OsDREB2C | Oryzasativa | AK108143 | None | Matskura et al. [19]. |
| OsDREB2E | Oryzasativa | None | Matskura et al. [19]. | |
| OsDREBL | Oryzasativa | AF494422 | Cold | Chen et al. [20]. |
| TaDREB1 | Triticumaestivum | AAL01124 | Cold, Dehydration, ABA | |
| WCB2 | Triticumaestivum | Cold, Drought | - | |
| WDREB2 | Triticumaestivum | BAD97369 | Drought, Salt, Cold,ABA | - |
| HvDRF1 | Hordeumvulgare | AY223807 | Drought,salt,ABA | - |
| HvDREB1 | Hordeumvulgare | DQ012941 | Drought, Salt, Cold | - |
| ZmDREB2A | Zea mays | AB218832 | Drought,Salt, cold, Heat | - |
| PgDREB2A | Pennisetumglucum | AAV90624 | Drought,Salt, Cold | - |
| SbDREB2 | Sorghum bicolour | ACA79910 | Drought | - |
| SiDREB2 | Setarica italic | HQ132744 | Drought, Salt | Lata et al. [21] |
| CaDREB-LP1 | Capaicumanum | AY496155 | Drought, Salt, Wounding | - |
| AhDREB1 | Artiplexhortensis | Salt | - | |
| GmDREBa | Glycine max | AY542886 | Cold, Drought,Salt | - |
| GmDREBb | Glycine max | AY296651 | Cold,drought, Salt | - |
| GmDREBc | Glycine max | AY244760 | Drought, Salt,ABA | - |
| GmDREB | Glycine max | AF514908 | Drought, Salt | - |
| GmDREB2 | Glycine max | ABB36645 | Drought, salt | - |
| PpDBF1 | Pshycomitrella patens | ABA43697 | Drought, Salt,Cold, ABA | - |
| PNDREB1 | Arachis hypogeal | FM955398 | Drought, Cold | - |
| CAP2 | Cicerarietinum | DQ321719 | Drought, Salt,ABA, Auxin | - |
| DvDREB2A | Dendrathema | EF633987 | Drought, Heat, ABA, Cold | Liu et al. [13]. |
| DmDREBa | DendrathemaXmorifolium | EF490996 | Cold, ABA | Yang et al. [14]. |
| DmDREBb | DendrathemaXmorifolium | EF487535 | Cold, ABA | Yang et al. [14]. |
| PeDREB2 | Populous euphratica | EF137176 | Drought, Salt, Cold | Chen et al. [20]. |
| SbDREB2A | Salicorniabrachiata | GU592205 | Drought, Salt, Heat | - |
Table 2.Response of DREB’s in different type of oxidative stresses in plants.
3.1 Transcription factor regulates cold induced expression
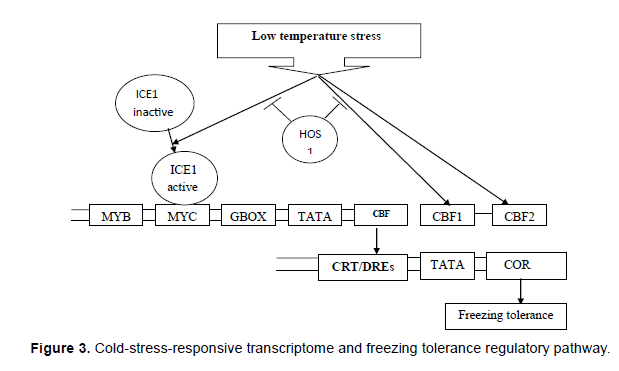
More than 100 genes are up regulated by cold stress. Because cold stress is clearly related to ABA responses and to osmotic stress, not all the genes up-regulated by cold stress necessarily need to be associated with cold tolerance. Many cold stress induced genes are activated by transcriptional activators called C-repeat binding factors (CBF1, CBF2, CBF3, also called DREB1b, DREB1c and DREB1a) [2]. CBF/DREB1 is involved in the coordinate transcriptional response of numerous cold and osmotic stress-regulated genes. CBF/DREB1b is unique in that it is specially induced by cold stress and not by osmotic or salinity stress, whereas the DRE- binding elements of the DREB2 type are induced only by osmotic and salinity stresses and not by cold. The expression of CBF1/DREB1b is controlled by a separate transcription factor, called ICE (inducer of CBF expression).
3.2 The ICE1 pathway regulates cold-stress responsive genes
The role of CBFs in cold-induced gene expression has been extensively studied. However, microarray analyses have indicated that multiple regulatory pathways are activated in response to cold, involving several other transcription factors [7,8]. The ICE1 (Inducer of CBF Expression 1)-CBF (C repeat binding protein) pathways is critical for the regulation of the cold-responsive transcriptome and acquired freezing tolerance. ICE1 is a constitutively expressed myc-like bHLH transcription factor, which is inactive under non-stress conditions. Low temperature stress presumably activates ICE1, which binds to myccis elements of the CBF3 promoter and induces CBF3 expression. CBFs binds to the CRT/DRE ciselements on cold-stress-responsive (COR) genes inducing their expression and leading to acquired freezing tolerance. A family of transcription factors known as C-Repeat binding factors (CBFs) or dehydration responsive element binding factors (DREBs) that control ABA-Independent expression of COR genes in response to cold stress has recently been identified (Figure 3).
3.3 Increases in CBF protein levels in response to cold and ABA
ABA signalling plays a vital role in plant stress responses. ABA inducible genes share the (C/T) ACGTGGC consensus, cis-acting ABA-response element (ABRE) in their promoter region [9]. Several ABRE-binding proteins, including rice TRAB and Arabidopsis AREB/ABF and AB15, which interact with ABRE and regulate gene expression, have been isolated (Hobo et al. 1999). Recently Abe et al. showed that the Arabidopsis MYB transcription factor. These three genes are inducible by cold. ABA levels rise transiently in response to low temperature. Over expression of CBF2 is sufficient to increase coldinduced gene expression and subsequently freezing tolerance [10]. On the contrary, over-expression of CBF2 is sufficient to increase cold-induced gene expression and subsequently freezing tolerance because of the common gene targets of the three CBF proteins [11].
Since CBF transcripts begin accumulating with in 15 min of plant exposure to cold, it has been proposed that there is a transcription factor already present in the cell at normal growth temperature that recognizes the CBF promoters and induced CBF expression upon exposure to cold stress [6].
4. Conclusion
The phytohormones ABA is produced by plants under cold and drought stress conditions and plays an important role in mediating stress tolerance. ABA, cold and osmotic stresses are known to induce expression of many of the same genes in Arabidopsis. Induction via this element involve the action of the CBF (DREB1; cold) and DREB2 (drought) families of TFs and their homolog’s [5,12-21]. Three CBFrelated genes encoding proteins that binds to the CRT/DRE. CBF4 (DREB1D), can be expressed in response to ABA have identified three CBF-related genes encoding proteins that bind to the CRT/DRE. CBF4 (DREB1D) can be expressed in response to ABA. The increase in CBF1-3 transcript levels report may have been more easily observed in the system. ABA may be able to potentiate cold-induced CBF signalling even if the ABA and cold stimuli do not occur concurrently.
References
- Shinozaki K, Yamaguchi-Shinozaki K. (1997). Gene expression and signal transduction in water-stress response. Plant Physiol: 327–334.
- Shinozaki K, Yamaguchi-Shinozaki K. (2000). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol.217–223.
- Chapman D. (1998). Phospholipase activity during plant growth anddevelopment and in response to environmental stress. Trends Plant Sci. 419-426.
- Ingram J, Bartel D. (1996). The molecular basis of dehydration tolerance in plants. Annu Rev Plant PhysiolPlant Mol Biol.377-403.
- Stockinger EJ.(1997). Arabidopsis thalianaCBF1 encodes an AP2 domain-containing transcriptional activator that binds to the Crepeat/ DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci U S A. 1035-1040.
- Gilmour SJ, Zarka DG, Stockinger EJ, et al. (1998). Low temperatureregulation of the ArabidopsisCBF family of AP2 transcriptionalactivators as an early step in cold-induced COR gene expression. Plant J. 433–442.
- Fowler S, Thomashow MF. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 1675–1690.
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, et al.(1998). Arabidopsis CBF1 overexpressioninduces COR genes and enhances freezing tolerance. Science. 104–106.
- Guiltinan MJ, Marcotte WR, Quatrano RS. (1990). A plant leucine zipper protein that recognizes an abscisic acid response element. Science. 267–271.
- Novillo F, Alonso JM, Ecker JR, et al. (2004). CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc Natl Acad Sci USA. 3985–3990.
- Gilmour SJ, Fowler SG, Thomashow MF. (2004). Arabidopsis transcriptional activators CBF1, CBF2 and CBF3 have matching functional activities. Plant Mol Biol. 767–781.
- Haake V, Cook D, Riechmann JL, et al. (2002). Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 639–648.
- Kasuga M, Liu Q, Miura S, et al. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. NatBiotechnol.287–291.
- Hsieh TH, Lee JT, Yang PT, et al. (2002). Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol. 1086–1094.
- Abe H, Urao T, Ito T, et al. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell: 63–78.
- Lee H, Xiong L, Gong Z, et al. (2001). The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING Rnger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes and Development.15: 912-924.
- Wang W, Vinocur B, Altman A. (2003). Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 1–14.
- Dubouzet JG, Sakuma Y, Ito Y, et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought, highsalt and coldresponsive gene expression.Plant J. 33: 751-63.
- Matsuura K, Hayashi N, Kuroda Y, et al. (2010). Improved development of mouse and human embryos using a tilting embryo culture system. Reprod Biomed Online.20: 358-364.
- Chen L, Lee D, Song Z, et al. (2004). Gene flow from cultivated rice (Oryza sativa) to its weedy and wild relatives. Ann Bot.93: 67–73.
- Lata C, Prasad M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. Journal of Experimental Botany. 4731–4748.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences