Identification, Structural and Evolutionary Analysis of Polyprenyl Synthase Gene Family in Tomato and Three Tobacco Varieties
F Li, YL Xu, CY Wei, MZ Wu, Z Wang, JF Zhang, R Wang, P Wang, J Yang, LF Jin
F Li1, YL Xu1, CY Wei2, MZ Wu1, Z Wang1, JF Zhang1, R Wang1, P Wang3, J Yang1, LF Jin1,*
1China Tobacco Gene Research Center, Zhengzhou Tobacco Research Institute, Zhengzhou, China
2Staff Development Institute of CNTC, Zhengzhou, China
3Tropical Crops Genetic Resources Institute, Chinese Academy of Tropical Agricultural Sciences, Danzhou, China.
Received date: August 04, 2016; Accepted date: October 14, 2016; Published date: October 21, 2016
Citation: Li F, Xu YL, Wei CY, et al. Identification, Structural and Evolutionary Analysis of Polyprenyl Synthase Gene Family in Tomato and Three Tobacco Varieties. Electronic J Biol, 12:4
Copyright: © 2016 Mbamalu ON, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Polyprenyl synthase gene family contained many genes which played key roles in terpenoids biosynthesis. In A. thaliana, 17 polyprenyl synthase genes were already identified which could be divided into 4 major groups as Geranylgeranyl Diphosphate Synthase (GGPPS), Farnesyl Diphosphate Synthase (FPS), Solanesyl Diphosphate Synthase (SPS) and Geranyl Diphosphate Synthase (GPS). In this article, bioinformatics analyses were performed to infer the putative polyprenyl synthase genes of tomato and three tobacco varieties based on whole genome databases. In total, 13 and 32 polyprenyl synthase genes were identified in whole genome of tomato and three tobacco varieties. Phylogenetic analyses showed that they were grouped into three major clades, gps and sps genes shared relative close relationship for one subclade grouping. Besides of these, 10 positive selection sites were identified along the sequences of Sub III clade which contained sps and gps genes. 2 of 10 positive selection sites were mapped onto 3-dimensionmal structures of tomato SPS and GPS sequences, respectively. One positive selection site was neighbor of conserved domain DDXXD, suggesting its important role played in polyprenyl synthase activities. Tomato GGPPS gene only has 1 intron, while other genes have multiple introns.
Keywords
Polyprenyl synthase; Geranylgeranyl diphosphate synthase; Farnesyl diphosphate synthase; Solanesyl diphosphate synthase; Geranyl diphosphate synthase; Evolutionary analysis.
Introduction
Available genomic data present an unprecedented opportunity for molecular evolution bias analyses [1]. For Solanaceae species, several whole genome sequenced articles have been published, such as tomato, pepper, potato, tobacco and relative field type species, which provided possibilities in clear clarification of gene family member evolution characteristics. Terpenoids, also called isoprenoids, are major metabolites of plants, which played important roles in crops quality, such as tomato, tobacco and others [2]. For terpenoids biosynthesis of plants, two major pathways have been published, named Mevalonic Acid Pathway (MVA) and Methylerythritol Phosphate Pathway (MEP), which catalyzed by large number of enzymes [3,4]. An important class of enzymes in plant terpenoids biosynthesis is polyprenyl synthase gene family, which contains PF00348 domain as revealed in Pfam database.
Based on Pfam database searching, PF00348 domain was found in many Arabidopsis thaliana peptide sequences, which have been identified as Solanesyl Diphosphate Synthase (SPS), Geranyl Diphosphate Synthase (GPS), Farnesyl Diphosphate Synthase (FPS) and geranylgeranyl diphosphate synthase (GGPPS), and all sequences were associated with terpenoids biosynthesis [5].
SPS was encoded by two isoforms in Arabidopsis thaliana, were AT1G78510 (SPS1) and AT1G17050 (SPS2) in whole genome database, which was regarded as the precursor of the side chains of both plastoquinone and ubiquinone [6]. Subcellular localization analyses showed that they are located in different organelles: SPS1 was located in ER while SPS2 was in plastid. So SPS1 and SPS2 were inferred to associate with the side chains of ubiquinone and plastoquinone, respectively [6].
GPS catalyzes the condensation of the key precursor of monoterpene biosynthesis. 2 genes were cloned and identified as coding sequences of Peppermint gps, which were named pMp13.18 and pMp23.10, respectively [7]. Gene expression analyses showed that only co-expression of two genes can yield detected prenyl transferase activity. In Arabidopsis thaliana, only one gene was identified as coding sequence of gps, whose gene ID was AT2G34630. This gene was characterized to be required for biosynthesis of gibberellins [8].
FPS catalyzes the synthesis of the major substrate used by cytosolic and mitochondrial branches [9]. Two fps genes were identified in whole genome of Arabidopsis thaliana, named fps1 and fps2 [10]. fps1 has a predominant role during most of the plant life cycle, and fps2 appears to have a major role in seeds and during the early stages of seedling developments [11]. A fps gene was cloned in tomato, labeled as Lefps1; molecular biology analyses showed that multiple fps isoforms were involved in tomato farnesyl pyrophosphate metabolism and that fps genes were mostly expressed in relation to cell division and enlargement [12].
GGPPS catalyzes the precursor for the biosynthesis of gibberelins, carotenoids, chlorophylls, isoprenoids quinones and geranyl greanylated proteins in plants. Several ggpps isoforms have been identified in whole genome of Arabidopsis thaliana. 5 Arabidopsis ggpps genes expressed in different organs of plants, and there would be specific pathways of GGPP production in each organelle. Whole genome identification showed that 12 ggpps genes were contained in Arabidopsis genome, and subcellular location and tissue-expression analyses suggested their sub functionalization in providing GGPP to specific tissues, developmental stages or metabolic pathways [13].
Above mentioned genes were important for terpenoids biosynthesis, while none of systemic analyses were performed to identify putative isoforms and clarify the molecular evolution and gene family expansion bias. In this paper, 45 polyprenyl synthase gene family members were identified in whole genomes of tomato and three tobacco cultivars, and showed that these genes were divided into three sub-lineages in phylogenetic tree. Besides, the molecular evolution characteristics of each sub-lineage were analyzed and 10 positive selection sites along the sequences of Sub III clade were identified.
2. Materials and Methods
2.1 Sequence retrieval and basic characteristics analyses
Gene models of tomato and three tobacco cultivars were downloaded from sol genomics network, and A.thaliana was obtained from TAIR 10. The data was processed as described [14]. 12 A. thaliana GGPPS genes, which have been identified previously, were used as queries for the further BLASTP analyses. BLAST searches, which used BLASTP as engine, were performed against the peptide datasets as described above with a rather high E value of 50 for coverage of some low similarity sequences. Besides of these, HMM searches of the protein datasets were also performed with the “trusted cut-off” established by Pfam databases [15]. The sequences of above two searches were merged; duplication loci were deleted, with a raw sequences file obtained.
The above mentioned raw sequence file was searched against Pfam_A library though pfam_scan. pl scripts download from Pfam website, with e_seq and e_dom set as 1e-3 and 1e-6, respectively. From the results, only the output items with PF00348 as hmm ID and with significance as 1 were selected, which generated a pool of locus identifiers. Then, the protein and coding sequences were retrieved based on the locus ID of this pool. For multiple sequences with alternative splicing, only the longest sequences were retained.
2.2 Phylogenetic tree reconstruction
Sequence alignments were performed using Probcons and PRANK, respectively, with default parameters, and the results were manually edited through MEGA 6.0 and saved as FASTA and nexus files for subsequent analysis [16-18]. Coding sequences alignments were used to reconstruct phylogenetic trees through Bayesian inference (BI) method through Mrbayes version 3.2.5, with 2,000,000 generations, 2 runs, 4 Markov chains and 0.1 temperature; split frequencies were checked after 2,000,000 generations, which turned out to be under 0.01. Tree files were visualized with Figtree software [19,20].
2.3 Molecular evolution analyses
CODEML program in PAML packages was used to clarify the molecular evolution characteristics of polyprenyl synthase gene family members through calculation of codon non-synonymous/synonymous rate ratio (ω=dN/dS). Consistent sites used in PAML software were produced in PAL2NAL online server with setting protein alignments and coding sequences alignments as reference [21]. Tree file of PAML was newick format produced by using figtree software. Site models, allows ω ratio to vary among sites, were used to identify positive selection sites and point out selection pressures distribution among different sites. Three pairs of LRT comparison were used in this analysis. The first pair including M0 (one ratio) and M3 (discrete) was performed to test variable ω ratios among sites; the second pair including M1a and M2a and the third pair including M7 and M8 were used to identify positive selection sites. The LRT comparison was decided to be significant if P-value was lower than 0.05. The branch site models, assuming that ω ratios were varied between foreground branches and background branches, were used to detect positive selection sites along special sequences. In the LRT, branch site model A is the alternative model, while the simpler null model is model A but with ω2=1 fixed. The significant LRT comparison (P-value<0.05) between Model A and Model A null indicate some sites of foreground branches evolved under positive selection pressures. In site model and branch site model analyses of this paper, the Posterior Probabilities (PP) of each potential positive selection amino acid sites were calculated by using Bayes Empirical Bayes (BEB) method and sites with PP values higher than 0.9 were considered as positive selection sites.
2.4 Three-dimensional modeling and gene structure analyses
Three-dimensional of target sequences were modelled in Swiss-model online server [22]. The templates of each sequence were searched and determined in known protein 3-dimensional structures database. The output files were saved as PDB format and then loaded into PyMOL file for display and edit [23]. Exon and UTR feature information was retrieved from GFF data of tomato genome and visualization of gene structures were carried out using GSDS 2.0.
3. Results and Discussion
In total, 13 and 32 polyprenyl synthase gene family members were identified in whole genomes of tomato and 3 tobacco varieties, respectively. Gene identification results in genomes of tobacco cultivars showed that K326 and TN90 11 genes were identified in varieties of K326 and TN 90, respectively, while 10 genes were retrieved in variety of BX [24]. The missing genes of BX may be due to poor assembly and annotation quality of the genome. 3 tobacco cultivars shared similar gene numbers with tomato. 17 Arabidopsis thaliana polyprenyl synthase genes, extracted from multiple literatures, were used as reference for gene identification in this study, whose roles were listed in Table 1.
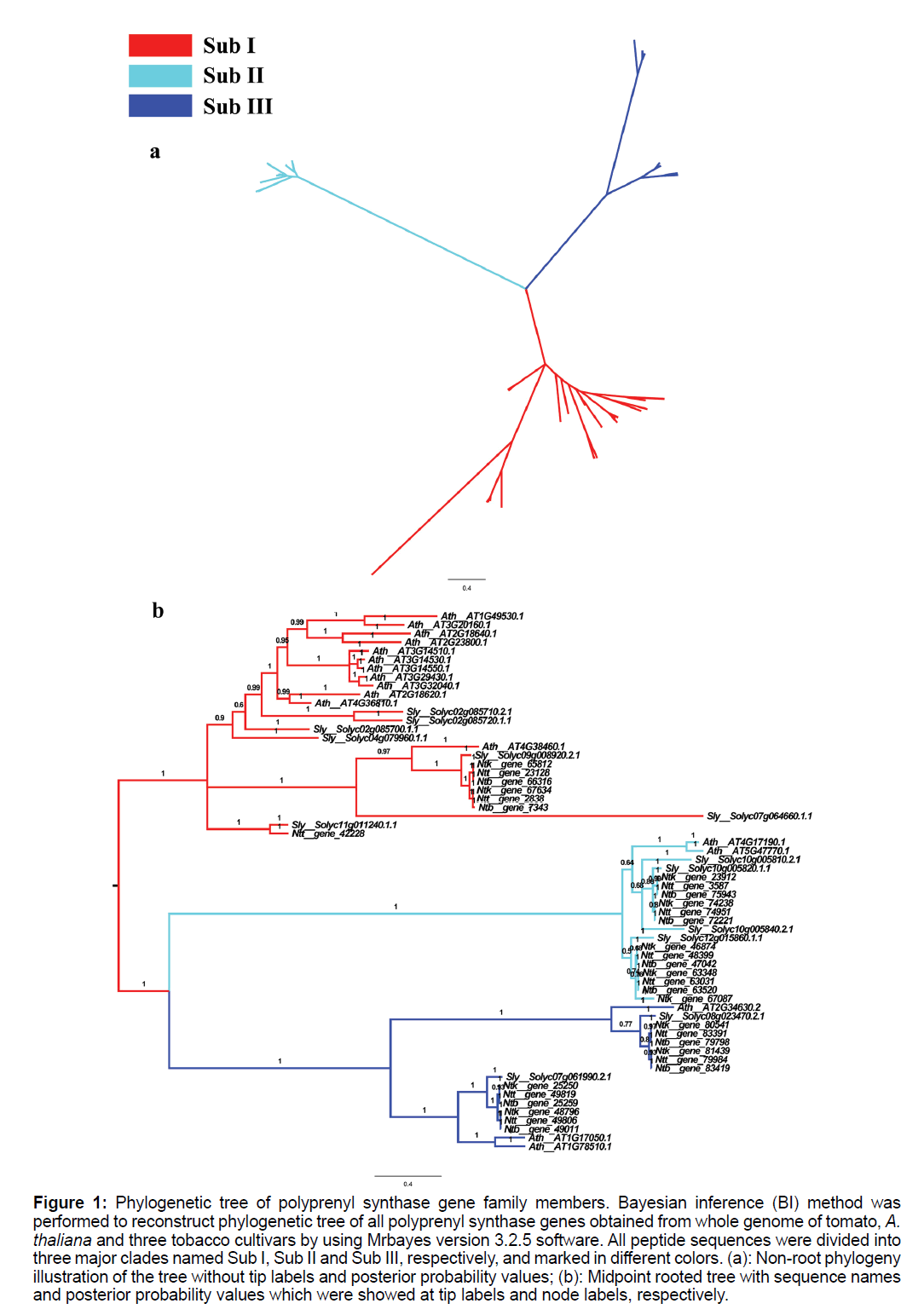
To understand the origin and evolutionary history of Solanaceae polyprenyl synthase genes, and to gain an insights into their molecular roles, the coding sequences of the 45 Solanaceae polyprenyl synthase gene models, together with 17 A. thaliana genes, were used to reconstruct Bayesian phylogenetic tree with WAG substitution model. Midpoint rooted Bayesian inference phylogenetic tree of all 62 polyprenyl synthase gene family members were displayed, which revealed three major clades for this family (Figure 1), labeled as Sub I, Sub II and Sub III, respectively. For A. thaliana, over 70% of genes fell into Sub I, while genes in Sub II and Sub III accounted for only 12-18% of total gene numbers. However, Solanaceae genes were partitioned into 3 major clades with similar number. Concerning the 17 A. thaliana polyprenyl synthase genes already characterized (Table 1), in Sub I, 12 ggpps genes were included; fps1 and fps2 were in Sub II; in Sub III, two isoforms of sps genes and one gps gene were included.
| ID | Function | Abbreviation | Literature |
|---|---|---|---|
| At1g78510.1 | Solanesyldiphosphate synthase | SPS1 | Jun et al. [6] |
| At1g17050.1 | SPS2 | Jun et al. [6] | |
| At2g34630.1 | Geranyldiphosphate synthase | GPS1 | Van Schieet al. [8]; Bouvieret al. [32] |
| At5g47770.1 | Farnesyldiphosphate synthase | FPS1 | Closaet al. [11] |
| At4g17190.1 | FPS2 | Closaet al. [11] | |
| At1g49530.1 | Geranylgeranyldiphosphate synthase | GGPPS1 | Willeet al. [30]; |
| At2g18620.1 | GGPPS2 | ||
| At2g18640.1 | GGPPS3 | ||
| At2g23800.1 | GGPPS4 | ||
| At3g14510.1 | GGPPS5 | ||
| At3g14530.1 | GGPPS6 | ||
| At3g14550.1 | GGPPS7 | ||
| At3g20160.1 | GGPPS8 | ||
| At3g29430.1 | GGPPS9 | ||
| At3g32040.1 | GGPPS10 | ||
| At4g36810.1 | GGPPS11 | ||
| At4g38460.1 | GGPPS12 | ||
Table 1. Functions and original literatures of 17 A. thaliana polyprenyl synthase genes used in this study.
Figure 1: Phylogenetic tree of polyprenyl synthase gene family members. Bayesian inference (BI) method was performed to reconstruct phylogenetic tree of all polyprenyl synthase genes obtained from whole genome of tomato, A. thaliana and three tobacco cultivars by using Mrbayes version 3.2.5 software. All peptide sequences were divided into three major clades named Sub I, Sub II and Sub III, respectively, and marked in different colors. (a): Non-root phylogeny illustration of the tree without tip labels and posterior probability values; (b): Midpoint rooted tree with sequence names and posterior probability values which were showed at tip labels and node labels, respectively.
In multiple alignments of all analyzed sequences (Figure S1), three conserved motifs previously reported were only identified in Sub I clade. In Sub II and Sub III, CXXXC and DDXXXXD were lost and two DDXXD motifs were identified.
3.1 ggpps genes in Solanaceae
Sub I contains 12 A. thaliana genes (Figure 1 and Table 1) that are known to encode GGPPS, which catalyzes the biosynthesis of GGPP, an intermediate in the biosynthesis of terpenoids. The enzyme is believed to be important in various primary and specialized metabolisms.
To test if the genes in this clade are also ggpps genes, deduced peptide sequences of the 14 Sub I genes were compared with peptide sequences in the following species: (1) a human ggpps gene (AAH05252.1); (2) genes from 3 plant species, which were already identified and characterized to be ggpps genes [Taxus canadensis (AAD16018.1), Capsicum annuum (CAA56554.1), Hevea brasiliensis (BAB60678.1) [25-27]. For CXXXC motif, both of cystine residues were replaced by serine and asparagine respectively in human ggpps gene; For DDXXD motif, the last aspartic acid residue was mutated as glutamic acid in all sequences of tobacco, and the common mutation was also identified in one sequence of tomato (Solyc09g008920.2.1) and A. thaliana (AT4G38460.1). In addition, this motif was lost in a tomato sequence labelled as Solyc07g064660.1.1. For DDXXXXD motif, all sequences contain this motif except losing in tomato sequence Solyc07g064660.1.1. In view of important functions of these motifs, this sequence (Solyc07g064660.1.1) was deleted in further analyses.
Overall, based on the results of multiple alignments and motif identification, the genes in Sub I were partitioned with well-characterized ggppss except Solyc07g064660.1.1, and thus labeled genes in this clade as ggpps genes.
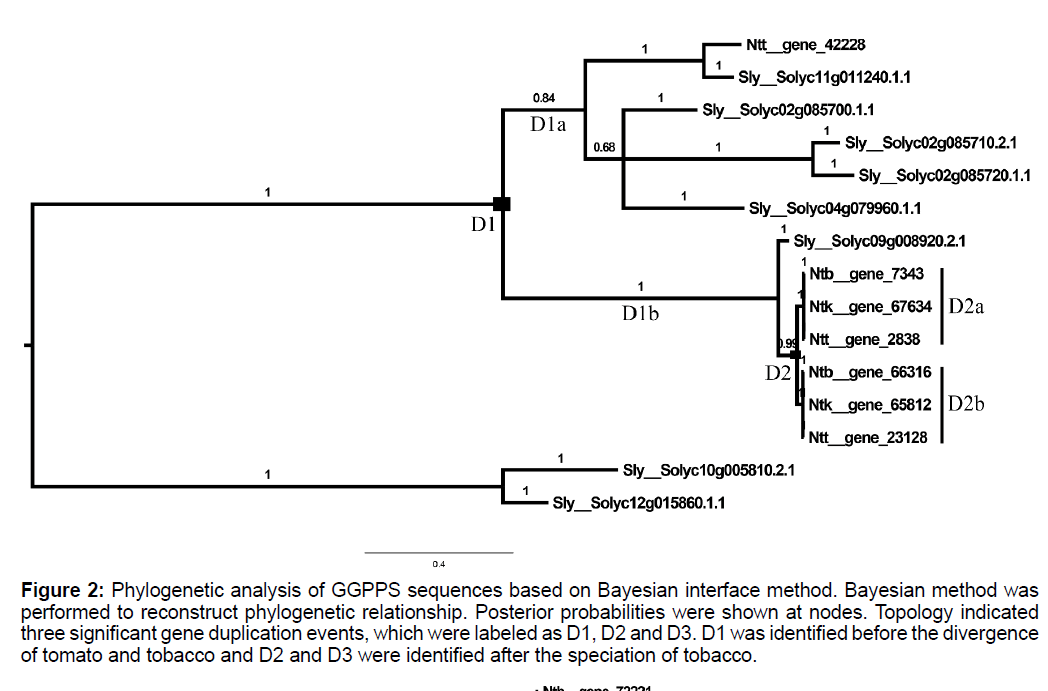
To shed light on evolutionary relationships of ggpps genes among tomato and 3 tobacco cultivars, Bayesian interface method was performed to reconstruct phylogenetic of these putative GGPPS with two tomato sequences of another clade (Solyc10g005810.2.1 and Solyc12g015860.1.1) as out group, with amino acid substitution model as WAG. Results indicated that two significant gene duplication events, which were labelled as D1 and D2 in Figure 2, were identified in the evolution of Solanaceae. Interestingly, the third aspartic acid site of DDXXD motif was mutated to glutamic acid (Figure S2) in D1b branch of Figure 1, while none of function changes about this mutation were reported. CODEML software in PAML package was used to clarify molecular evolution characteristics of Solanaceae ggpps genes using site model analysis, as shown in Table S1. Model M0 and Model M3 were conducted to test whether there was rate heterogeneity among amino acid sites and the significant LRT results among these two models indicated that the omega values of different sites were distributed as discrete. The detailed distribution characteristics of omega values were shown in Figure S3, which showed that all sites of GGPPS peptide sequences evolved under purifying selection pressures, and N-terminal evolved under more relaxed selection constraint. None of sites were identified to evolve under positive selection pressures for non-significant LRT results of M1a vs. M2a and M7 vs. M8.
3.2 fps genes in Solanaceae
FPS catalyzes Farnesyl Diphosphate (FPP), which serves as major substrate used by cytosolic and mitochondrial branches of the isoprenoid pathway. In A. thaliana, FPS was encoded by two genes which named fps1 and fps2, respectively. Knockout of both of genes (fps1 and fps2) was lethal, while no major developmental and metabolic defects were observed in fps1 and fps2 single knockout mutants. The gene (Accession: GQ420346.1) encoding FPS was cloned from a cDNA library of Artemisia annua, whose heterogeneous expression of Escherichia coli showed enzyme activity for A. annua FPS (AanFPS) in vitro [28].
In this article, both of A. thaliana fps genes were contained in Sub II clade (Figure 1), which formed a clade distinct to other clades of polyprenyl synthase genes. Three Solanaceae species analyzed in this paper were represented in this clade. For each species, there were 4 copies in this clade.
Six aspartates in two conserved domains (DDXXD) were reported in many articles, which showed that these domains were important for enzyme activity of FPS. Multiple alignments indicated that both of domains were in all sequences except two of tomato sequences (Solyc10g005840.2.1 and Solyc10g005820.1.1) and one tobacco K326 sequence (gene_67087) (Figure S1). Both sequences were deleted in further analyses.
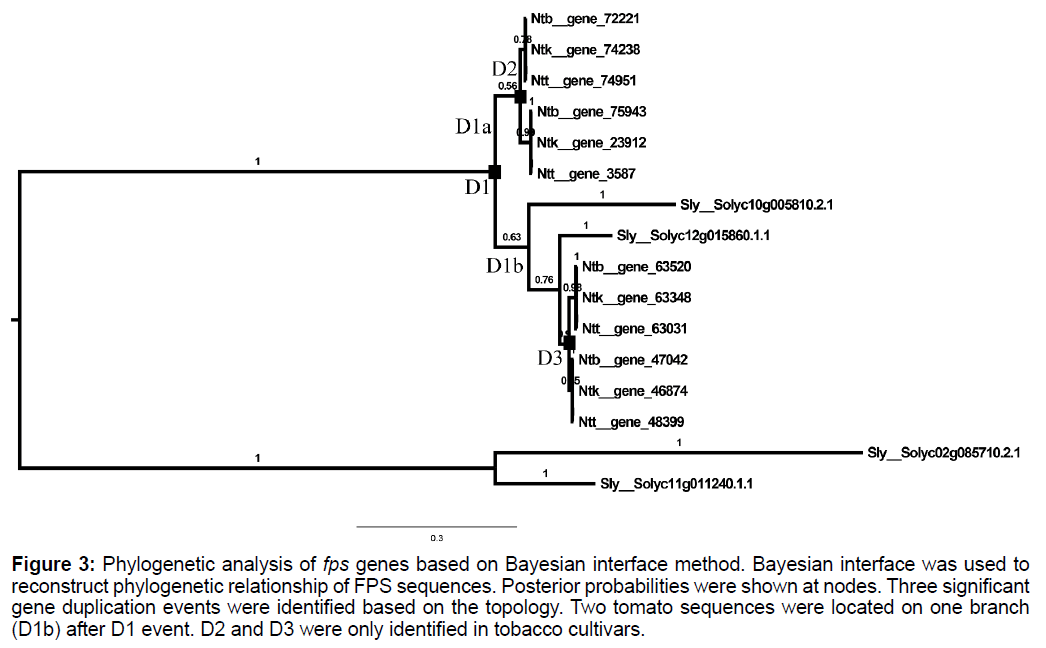
Using 2 tomato GGPPS sequences (Solyc02g085710.2.1 and Solyc11g011240.1.1) as out group, reconstruction of phylogenetic tree of FPS peptide sequences were performed with amino acid substitution model WAG. Results showed that sequences of Sub II clade were divided into two specific clades (D1a and D1b in Figure 3). No tomato sequences were identified in D1a clade. Two independent gene duplication events were identified in Nicotiana genus which was labelled as D2 and D3 (Figure 3).
Figure 2: Phylogenetic analysis of GGPPS sequences based on Bayesian interface method. Bayesian method was performed to reconstruct phylogenetic relationship. Posterior probabilities were shown at nodes. Topology indicated three significant gene duplication events, which were labeled as D1, D2 and D3. D1 was identified before the divergence of tomato and tobacco and D2 and D3 were identified after the speciation of tobacco.
Figure 3: Phylogenetic analysis of fps genes based on Bayesian interface method. Bayesian interface was used to reconstruct phylogenetic relationship of FPS sequences. Posterior probabilities were shown at nodes. Three significant gene duplication events were identified based on the topology. Two tomato sequences were located on one branch (D1b) after D1 event. D2 and D3 were only identified in tobacco cultivars.
Molecular evolution analyses were performed to clarify evolutionary bias of Solanaceae fps genes through site-model in CODEML software in PAML package. As shown in Table S2, significant LRT between M0 and M3 indicated that the omega values among different amino acid sites were distributed as discrete. The distribution characteristics of omega values among sites were shown as histogram in Figure S4, which showed the sites with relax selection constraint (ω1=0.65470 in Table S2) distribute evenly throughout the sequences. None of sites evolved under positive selection pressures were detected for non-significant LRT results of M1a vs. M2a and M7 vs. M8.
3.3 gps and sps genes in Solanaceae
GPS catalyzes the condensation of dimethylallyl diphosphate and isopentenyl diphosphate to form geranyl diphosphate, which was the key precursor for monoterpene biosynthesis. In A. thaliana, GPS was encoded by only one gene, whose locus ID was AT2G34630.1 (Table 1). FPS, which plays key roles in isoprenoid biosynthesis, catalyzes the synthesis of farnesyl diphosphate from isopentenyl diphosphate and dimethyl allyl diphosphate. In A. thaliana, two gene copies of fps were previously identified (AT5G47770.1 and AT4g17190.1).
In Figure 1, A. thaliana FPS and GPS sequences were clustered in two subclades of Sub III clade, which indicated that the two genes shared closer relationship with each other than others. Besides of these, many genes in Solanaceae were identified in Sub III with clade divergence. Conserved motif identification showed that only DDXXD was contained in Solanaceae GPS and FPS peptides (Figure 1).
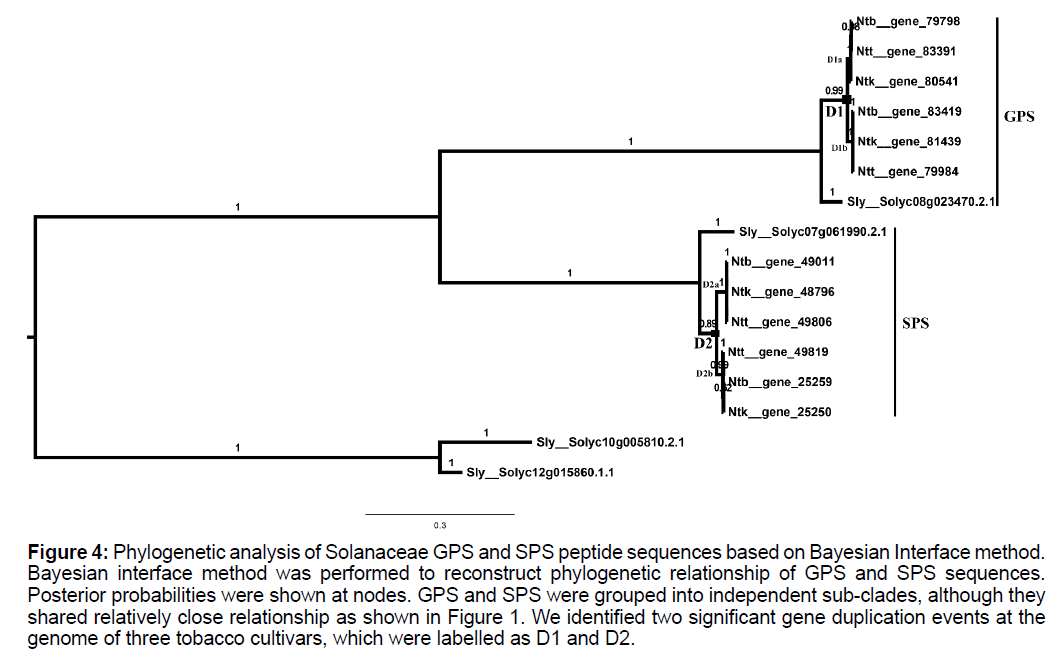
Phylogenetic topology of Solanaceae gps and sps genes was clarified based on Bayesian interface method. Results showed that SPS and GPS sequences of Solanaceae were split into two specific clades, which were consistent with results as shown in Figure 1. Two gene duplication events for these genes in three tobacco cultivars were identified in FPS and GPS clades, respectively, which were labelled as D2 and D3, respectively, in Figure 4.
Six site-models of CODEML software were used to analyze molecular evolution characteristics of Solanaceae fps and gps genes. The statistics results of molecular evolution analysis (Table S3) showed that several amino acid sites of target sequences evolved under discrete distribution omega values for Model M3, which significantly differed with Model M0. The omega value distribution histogram (Figure S5) showed that N-terminal evolved under more relaxed selection constraint than others, with some omega values above 1.
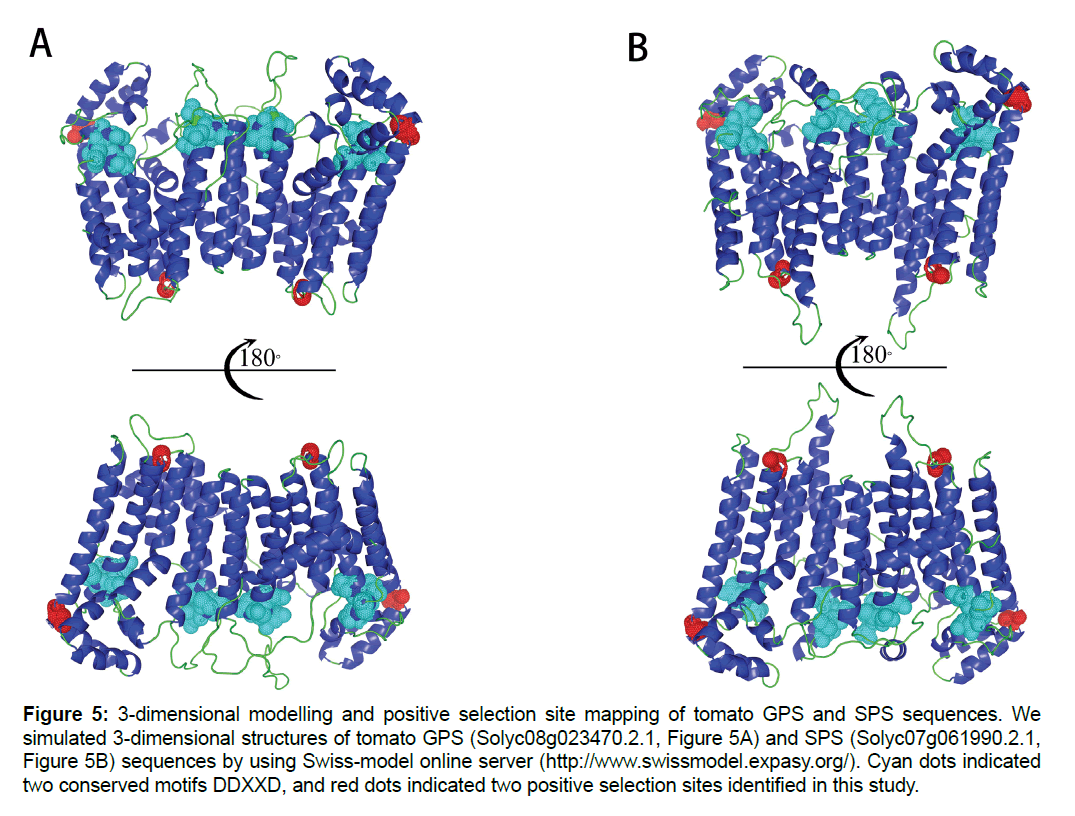
In positive selection site identification, the LRT statistics results showed non-significant comparison results among Model M2a and M1a, while very significant results were identified in LRT comparison between Model M7 and M8. Besides, BEB analysis showed that the omega values of 10 sites under Model M8 were higher than 1. Based on Yang [31], these 10 sites were positive selection sites. In order to clarify the putative functions of these sites, the sites were mapped onto 3-dimensional models of two tomato sequences which were located in two specific lineages of Sub III clade (D1a and D1b). Results showed that both of sites located on helix structure of protein. One of site (258D) was adjacent to conserved motif DDXXD, while another one (343T) was far away from these two motifs (Figure 5).
3.4 Gene structures of tomato polyprenyl synthase gene
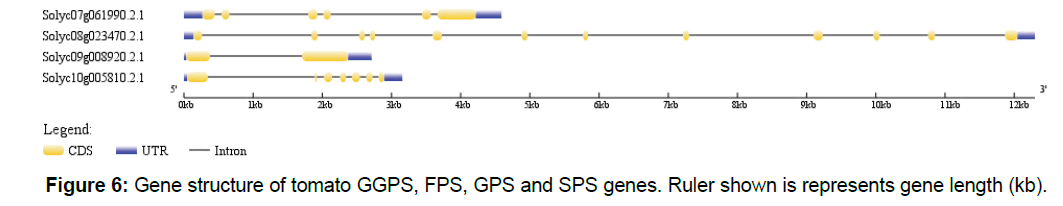
We have shown that genes in polyprenyl synthase family were split three major clades, which include four genes, and our results showed different evolutionary pressures for different clades. Besides CDS, genes in eukaryotes usually contain UTR and introns. We retrieved gene structure information from GFF3 file of tomato genome sequencing data for GPPS, GPS, FPS and SPPS genes. As shown in Figure 6, tomato GGPS gene (Solyc09g008920.2.1) only contains 1 intron, while FPS (Solyc10g005810.2.1), GPS (Solyc08g023470.2.1) and SPS (Solyc07g061990.2.1) genes contain multiple introns. GPS has 11 introns, SPS contains 5 introns, and FPS has 6 introns.
Figure 4:Phylogenetic analysis of Solanaceae GPS and SPS peptide sequences based on Bayesian Interface method. Bayesian interface method was performed to reconstruct phylogenetic relationship of GPS and SPS sequences. Posterior probabilities were shown at nodes. GPS and SPS were grouped into independent sub-clades, although they shared relatively close relationship as shown in Figure 1. We identified two significant gene duplication events at the genome of three tobacco cultivars, which were labelled as D1 and D2.
Figure 5:3-dimensional modelling and positive selection site mapping of tomato GPS and SPS sequences. We simulated 3-dimensional structures of tomato GPS (Solyc08g023470.2.1, Figure 5A) and SPS (Solyc07g061990.2.1, Figure 5B) sequences by using Swiss-model online server (https://www.swissmodel.expasy.org/). Cyan dots indicated two conserved motifs DDXXD, and red dots indicated two positive selection sites identified in this study.
4. Discussion
Genes in polyprenyl synthase (pfam: PF00348) gene family are important for biosynthesis of terpenoids based on the studies of functioned PF00348 genes in multiple species. In this paper, we identified 32 putative polyprenyl synthase genes from whole genome databases of tomato and three tobacco varieties (TN90, K326 and BX) by using bioinformatics tools. Phylogenetic tree analysis showed that all genes were divided into three major clades, and four types of genes have been inferred as the reference of all functioned A. thaliana genes. Results showed that putative genes in polyprenyl synthase gene family included ggpps, fps, sps and gps, respectively. In view of phylogenetic relationships of four gene types, we found that gps and sps genes shared close relationship, which might be the result of recent gene duplication and divergence from a common ancestral sequence.
For ggpps genes, showed that 5 A. thaliana ggpps genes were located in three different subcellular compartments and were expressed in multiple tissues, which indicated that these genes were not redundant sequences and might involve in different pathways in plants [29]. Previous reports identified 12 ggpps genes in whole genome of A. thaliana, and 10 of 12 sequences were found as functional proteins [30]. In phylogenetic analysis of this paper (Figure 1), we showed that all genes of GGPPS1 to GGPPS 11 were grouped as a single clade, which indicated that these genes may diverge from recent genome duplication events. However, GGPPS12, which has no GGPPS function, was grouped in another clade of Sub I and shared close relationship with some genes in tomato and tobacco. Therefore, we inferred that some proteins without GGPPS function were also contained in GGPPS subfamilies of tomato and tobacco [13,30].
Two differently expressed fps genes have been identified in whole genome of A. thaliana [10]. Phylogenetic analysis (Figure 1) showed that these two genes were grouped as a single clade of Sub II, which was consistent with the high sequence similarity previously reported [10]. One significant gene duplication event divided all fps like genes of Nicotiana genus into two subclades (Figures 1 and 3). However, fps genes of tomato were absent in one of the clade (D1a of Figure 3). For tomato, the absence could again be due to the genome sequencing and assembly gaps; it is also conceivable that no gene duplication event took place for tomato fps subfamily. Two sps and one gps genes, which shared close relationship in phylogenetic analysis, were identified in whole genome of A. thaliana [10]. We also identified some sequences of tomato and three tobacco varieties, which were grouped into A. thaliana gps and sps clades. We inferred that the close relationship may be due to recent gene duplication and divergence from common ancestral sequence (gps and sps).
Molecular evolution analyses were performed in this paper to clarify the evolution characteristics of each subfamily. We could not find positive selection sites in GGPPS and FPS groups, which indicated that these genes may be important for biological processes and thus evolved conservatively. However, we detected two sites evolved under positive selection pressures in groups which contained sps and gps genes. Positive selection sites identification indicated that some available mutations were retained in the evolution pathway, and these sites may be important for functions of protein [31,32]. We also found one of positive selection sites was neighbor to conserved domain of protein, suggesting its possibly important roles played in GGPPS and GPS enzymatic functioning.
In summary, we identified putative polyprenyl synthase genes from whole genome of tomato and three tobacco varieties by using in silico methods.
Phylogenetic and multiple alignments analysis divided all polyprenyl synthase genes into three major clades and four subfamilies. Besides we preformed molecular evolution analysis to highlight evolution characteristics of these genes. Based on this paper, we provide data which should be important for further analysis of terpenoids biosynthesis in Solanaceae.
5. Acknowledgement
The authors thank the Zhengzhou Tobacco Research Institute for support offered during the preliminary study. This work was supported by the Natural Science Foundation of China (31000137) the Science Project of the Zhengzhou Tobacco Research Institute (902012CZ0340).
References
- Hamilton JP, Robin Buell C. (2012). Advances in plant genome sequencing. Plant J.70:177-190.
- Langenheim JH. (1994). Higher plant terpenoids: Aphytocentric overview of their ecological roles. J. Chem. Ecol. 20:1223-1280.
- McCaskill D, Croteau R. (1993). Procedures for the isolation and quantification of the intermediates of the mevalonic acid pathway. Anal Biochem. 215:142-149.
- Hoeffler JF, Tritsch D, Grosdemange-Billiard C, et al. (2002). Isoprenoid biosynthesis via the methylerythritol phosphate pathway. Mechanistic investigations of the 1-deoxy-D-xylulose 5-phosphate reductoisomerase. Eur. J. Biochem.269:4446-4457.
- Finn RD, Bateman A, Clements J, et al. (2014). Pfam: The protein families database. Nucleic Acids Res.42:D222-230.
- Jun L, Saiki R, Tatsumi K, et al. (2004). Identification and subcellular localization of two solanesyldiphosphate synthases from Arabidopsis thaliana. Plant Cell Physiol.45:1882-1888.
- Burke CC, Wildung MR, Croteau R. (1999). Geranyldiphosphate synthase: cloning, expression and characterization of this prenyltransferase as a heterodimer. ProcNatlAcad Sci.96:13062-13067.
- Van Schie CC, Ament K, Schmidt A, et al. (2007). Geranyldiphosphate synthase is required for biosynthesis of gibberellins. Plant J.52:752-762.
- Arró M, Manzano D and Ferrer A. (2014). Farnesyldiphosphate synthase assay. Methods Mol Biol.1153:41-53.
- Cunillera N, Arró M, Delourme D, et al. (1996). Arabidopsis thaliana contains two differentially expressed farnesyl-diphosphate synthase genes. J Biol Chem.271:7774-7780.
- Closa M, Vranová E, Bortolotti C, et al. (2010). The Arabidopsis thaliana FPP synthase isozymes have overlapping and specific functions in isoprenoid biosynthesis and complete loss of FPP synthase activity causes early developmental arrest. Plant J. 63:512-525.
- Gaffe J, Bru J-P, Causse M, et al. (2000). LEFPS1, a tomato farnesyl pyrophosphate gene highly expressed during early fruit development. Plant Physiol.123:1351-1362.
- Beck G, Coman D, Herren E, et al. (2013). Characterization of the GGPP synthase gene family in Arabidopsis thaliana. Plant Mol Biol.82:393-416.
- Wang P, Wang Z, Dou Y, et al. (2013). Genome-wide identification and analysis of membrane-bound O-acyltransferase. (MBOAT) gene family in plants. Planta.238:907-922.
- Punta M, Coggill PC, Eberhardt RY, et al. (2011). The Pfam protein families database. Nucleic Acids Res. 40:D290-301.
- Do CB, Mahabhashyam MS, Brudno M. (2005). ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res.15:330-340.
- Löytynoja A, Goldman N. (2010). webPRANK: A phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics. 11:579.
- Tamura K, Stecher G, Peterson D, et al. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. MolBiolEvol.30:2725-2729.
- Huelsenbeck JP, Ronquist F, Nielsen R, et al.(2001). Bayesian inference of phylogeny and its impact on evolutionary biology. Science.294:2310-2314.
- Ronquist F, Teslenko M, van der Mark P, et al. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol.61:539-542.
- Suyama M, Torrents D, Bork P. (2006). PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res.34:W609-W612.
- Biasini M, Bienert S, Waterhouse A, et al. (2014). SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res.42:W252-258.
- DeLano WL. (2002). The PyMOL molecular graphics system. DeLano ScientiÃÆïÃâìÃâÃÂc, San Carlos.
- Sierro N, Battey JN, Ouadi S, et al. (2014). The tobacco genome sequence and its comparison with those of tomato and potato. Nature Communications. 5.
- Kainou T, Kawamura K, Tanaka K, et al. (1999). Identification of the GGPS1 genes encoding geranylgeranyldiphosphate synthases from mouse and human. BiochimBiophysActa. 1437:333-340.
- Hefner J, Ketchum RE,Croteau R. (1998). Cloning and functional expression of a cDNAencoding geranylgeranyldiphosphate synthase fromTaxuscanadensisand assessment of the role of this prenyltransferasein cells induced for taxol production. ArchBiochemBiophys.360:62-74.
- Takaya A, Zhang YW, Asawatreratanakul K, et al. (2003). Cloning, expression and characterization of a functional cDNA clone encoding geranylgeranyldiphosphate synthase of Heveabrasiliensis. BiochimBiophysActa. 1625:214-220.
- Matsushita Y, Kang W, Charlwood BV. (1996). Cloning and analysis of a cDNA encoding farnesyldiphosphate synthase from Artemisia annua. Gene.172:207-209.
- Okada K, Saito T, Nakagawa T, et al. (2000). Five geranylgeranyldiphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol.122:1045-1056.
- Wille A, Zimmermann P, Vranová E, et al. (2004). Sparse graphical Gaussian modeling of the isoprenoid gene network in Arabidopsis thaliana. Genome Biol.5:R92.
- Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. MolBiolEvol.24:1586-1591.
- Bouvier F, Suire C, d'HarlingueA, et al. (2000). Molecular cloning of geranyldiphosphate synthase and compartmentation of monoterpene synthesis in plant cells. Plant J. 24:241-252.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences