Identification of Potential Drug for Dengue Hemorrhagic Fever by Network-Based Drug Reprofiling Approach
Praveenkumar KS*, Gnanasoundari Sekar, Mallady vashnavi Sri Kavya, Wan Suriana Wan Ab Rahman, Wan Nazatul Shima Shahidan and Gokulakannan Venkatesan
Published Date: 2022-07-02Praveenkumar KS1*, Gnanasoundari Sekar2, Mallady vashnavi Sri Kavya3, Wan Suriana Wan Ab Rahman4, Wan Nazatul Shima Shahidan4 and Gokulakannan Venkatesan4
1Department of Clinical Science, Narayana Institute of Cardiac Sciences, Bangalore, India
2Department of Biotechnology, Anna University BIT-Campus, Tiruchirappalli, India
3Department of Clinical Science, Osmania University, Hyderabad, India
4Department of Dental Sciences, Universiti Sains Malaysia (USM), Kelantan, Malaysia
- *Corresponding Author:
- Praveenkumar KS
Department of Clinical Science,
Narayana Institute of Cardiac Sciences, Bangalore,
India
E-mail:gomanpk33@gmail.com
Received date: April 20, 2022, Manuscript No. IPEJBIO-22-13259; Editor assigned date: April 25, 2022, PreQC No. IPEJBIO-22-13259 (PQ); Reviewed date: May 10, 2022, QC No. IPEJBIO-22-13259; Revised date: June 20, 2022, Manuscript No. IPEJBIO-22-13259 (R); Published date: July 02, 2022, DOI: 10.36648/1860-3122.18.9.041
Citation: Praveenkumar KS, Sekar G, Sri Kavya MV, Rahman WSWA, Shahidan WNS, et al. (2022) Identification of Potential Drug for Dengue Hemorrhagic Fever by Network-Based Drug Reprofiling Approach. Electronic J Biol, 18(9): 1-11
Abstract
Dengue fever can progress to Dengue Hemorrhagic Fever (DHF), which is a more serious and occasionally fatal form of the disease. The patient may acquire warning indications of serious disease about the time the fever begins to reduce (typically 3–7 days following symptom onset), and there are currently no effective antivirals available. Drug repurposing is emerging as a novel drug discovery process for rapidly developing effective DHF therapies. Through network pharmacology modeling, several FDA approved medications have already been researched for various viral outbreaks, by analyzing the interactions between virus-host gene interactions and therapeutic targets in the human genome network, a total of 45 repurposable medicines were discovered. Hub network analysis of host virus drugs association hypothesized that aspirin, captopril, rilonacept are efficient in the treatment of DHF, and gene enrichment analysis supports the findings. The human interactive contains the genes PTGS2, ACE, and F2, documented to have a role in the pathogenesis of disease progression of DHF, and our analysis of most of the drugs targeting these genes. As a result, genes targeting medications play a significant part in limiting the condition's advancement.
Keywords
Dengue hemorrhagic fever; Drug reprofiling; Network pharmacology; Network medicine
Introduction
Dengue fever, also known as "break bone fever," is characterized by an acute prevalence of severe fever three to fourteen days after being bitten by an infected mosquito. migraine, retro-orbital pain, myalgia’s, muscle aches, hemolytic anemia signs, rash, and a low white blood cell count are only a few of the symptoms [1]. Dengue Hemorrhagic Fever (DHF), a severe and sometimes fatal manifestation of the disease, affects certain dengue fever patients. The patient may acquire warning signs of serious disease about the period the fever began to diminish (typically 3–7 days following symptom onset). Severe abdominal discomfort, continuous vomiting, a significant change in temperature (from fever to hypothermia), hemorrhagic manifestations, or a change in mental status are indeed warning indicators (irritability, confusion, or obtundation). Restlessness, chilly clammy skin, a rapid weak pulse, and a narrowing of the pulse pressure (systolic blood pressure diastolic blood pressure) are all early indications of shock. Patients with dengue fever should be recommended to return to the hospital if any of these symptoms appear [2].
According to one estimate, 390 million dengue virus infections occur each year (95 percent credible interval 284–528 million), with 96 million (67–136 million) showing clinical symptoms (of any severity) [3]. According to dengue, 3.9 billion individuals are at risk of contracting the virus. Although there is a risk of infection in 129 nations, Asia bears 70% of the real burden. Over the last two decades, the number of dengue cases reported to WHO has increased by more than 8 times, from 505,430 cases in 2000 to over 2.4 million in 2010, and 5.2 million in 2019. Between 2000 and 2015, the number of deaths reported grew from 960 to 2032. This worrying rise in case numbers can be explained in part by a shift in national practices for recording and reporting dengue fever to health ministries and the World Health Organization. However, it also symbolizes the government's acknowledgment of the problem, and hence the need to disclose the prevalence of dengue fever 4As a result, while the complete global burden of the disease remains unknown, the observed growth only takes us closer to a more precise estimate of the full extent of the burden.
Dengue fever increased in Bangladesh, Brazil, the Cook Islands, Ecuador, India, Indonesia, the Maldives, Mauritania, Mayotte (Fr), Nepal, Singapore, Sri Lanka, Sudan, Thailand, Timor-Leste, and Yemen in 2020. Dengue fever is still a problem in Brazil, the Cook Islands, Colombia, Fiji, Kenya, Paraguay, Peru, and Reunion Island in 20215. The COVID-19 epidemic is putting enormous strain on healthcare and management systems all across the world. During this critical period, WHO has stressed the significance of maintaining efforts to prevent, identify, and treat vector-borne diseases such as dengue fever and other arboviral infections, as case numbers rise in various countries, putting urban people at risk for both diseases [4-6].
Recent systems biology developments suggest a unique testable hypothesis for systematic drug repositioning [7,8]. The time spent on R and D is greatly decreased when compared to traditional drug development programs. The typical strategy takes 10-16 years to develop a new treatment. A medication repositioning plan costs $1.6 billion to create, but a typical strategy costs $12 billion. The identification of new targets and illness proteins has been made possible by rapid advances in genomic, proteomic, structural, functional, and systems investigations of existing targets and other disease proteins. In this study, we provide an embedded medicine platform that uses a network based method to quantify the association of DHF with human host interaction, and then we examine the efficacy of existing FDA approved medications as potential repurposable associations with DHF host genes. To discover and prioritize existing pharmacological targets in the DHF pathway, FDA approved medications are chosen from a clinical registry database.
Materials and Methods
Building the dengue hemorrhagic fever human interactive
Based on our literature similarity searches and database similarity searches found out more than 588 dengue hemorrhagic targeting human (host) genes based similarity hit to score 50 we sort out 5 host interacting dengue hemorrhagic fever genes reported experimental evidence for interactions between human proteins and dengue virus proteins based on high throughput yeast two hybrid screening methods Recently, Dey and Mukhopadhyay reported the development of denvInt, a database of manually curated experimental data of dengue protein and host protein interactions. We merged the data of published references from denvInt and used it in our analysis along with the dengue host interactive data from the recent investigations [9-11].
Human (host) gene dengue hemorrhagic fever gene interactive
The key host genes involved in dengue hemorrhagic fever were identified from the gene card database using the search terms “dengue hemorrhagic fever; dengue hemorrhagic fever interacting human genes”. Gene card is a searchable, integrative database that provides comprehensive, user friendly information on all annotated and predicted human genes. The knowledge base automatically integrates gene centric data from ~150 web sources, including genomic, transcriptomic, proteomic, genetic, clinical and functional information as of January 13, 2022 gene card comprises 326,787 genes are available in which 18,870 disease genes and 500 hot genes [12].The functions genes identified from gene card and related literature were collected and presented in supplementary file 1. The PPI network was built with cytoscape 3.9.0 v and Gephi 0.9.2 v 13 software.
Drug targets (human genes) interactive
We collect 87 FDA approved antiviral and anti-dengue hemorrhagic fever drugs from the Therapeutic Target Database (TTD) compare them with the results of the drug bank database and identified drug targets and formulated them as a dataset its available in supplementary file 1. We visualized it using cytoscape [13-16]. Nodes in networks represent antiviral drugs or anti-dengue hemorrhagic fever drugs and the nodes of the network represent drugs targeting human genes [17].
Building the drug to human interactive
A network pharmacological based host dengue hemorrhagic fever-antiviral-anti dengue hemorrhagic fever drugs interactive was constructed by assembling the host dengue hemorrhagic fever interacted proteins with or without antiviral, anti-dengue hemorrhagic fever drugs. The PPI network was built with Gephi 0.9.2 v and cytoscape 3.9.0 v software. Each node in the constructed PPI network indicates a host gene and an edge indicates an interacting drug target [18].
Network hub gene identification
Highly connected nodes (hubs) in biological networks are topologically important to the structure of the network and have also been shown to be preferentially associated with a range of phenotypes of interest. Hub genes can be identified using the Contextual Hub Analysis Tool (CHAT) plug-in cytoscape 3.9.0 v which enables users to easily construct and visualize a network of interactions from a gene list of interest [19].
Network betweenness centrality analysis
Betweenness is a centrality measure of a vertex within a network; vertices that have a high probability to occur on a randomly chosen shortest path between two randomly chosen vertices have a high betweenness. The betweenness of a vertex α in a graph G: =(A,B) with A vertices and B edges [20]. For each pair of vertices (s,l), compute the shortest paths between them, For each pair of vertices (s,l), determine the fraction of shortest paths that pass through the vertex in question (here, vertex A). Sum this fraction over all pairs of vertices (sl). The degree Cb of node b is calculated as follows.
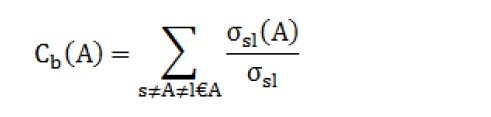
Were, ( A) is the number of shortest paths from s to l that pass through a vertex A [21].
In a connected graph, the normalized closeness centrality (or closeness) of a node is the average length of the shortest path between the node and all other nodes in the graph [22]. Thus, the more central a node is, the closer it is to all other nodes.

Were d (a,b) being the distance between vertices a and b
Functional enrichment analysis for genes and drugs
Functional enrichment analysis is a method to determine classes of genes or drugs that are over-represented in a large group of genes or drugs and may have relations with disease phenotypes. This approach uses statistical methods to determine significantly enriched groups of genes [23]. The biological relevance and functional pathways of our datasets were revealed by enriching the semantic similarities of the pathway, tissue. All functional enrichment analyses were performed using the enrichment platform as additional evidence for drug repurposing. The enrich is a comprehensive gene enrichment analysis platform that comprises 382,208 terms from 192 libraries. The combined score is described as
c=log (p).z
Where c=the combined score, p=Fisher exact test p-value, and z=z-score for deviation from expected rank.
Results
Human (Host) dengue hemorrhagic fever (viral) gene interactive
We constructed a host DHF interactive consisting of 59 interacting genes with 60 nodes and 59 edges (Supplementary Figure 1a). Based on Kegg pathway enrichment analysis indicates genes involved in the AGE-RAGE signaling pathway in diabetic complications enriched (P=3.01E-32) the most which indicate patients with DHF condition have a higher chance of poor blood sugar management while in the pathogenesis, AGE/RAGE signaling has been shown to increase oxidative stress to promote diabetes mediated vascular calcification through activation of Nox-1 and decreased expression of SOD-1 and chagas disease (P=4.65E-32)and influenza A pathways are typically enriched(P=6.17E-31) [24]. Compare to Kegg pathway analysis reactive pathway analysis indicates immune system (P=3.93E-28) and cytokine signaling in immune system (P=7.06E-27) are enriched which indicates DHF hijack human immune system associated gene pathway the most, gene set in which immune system (1.12E-61) bronchoalveolar lavage (2.85E-50) tissues are enriched the most (Supplementary Figure 1b).
Host-viral-antiviral drugs target interactive
A host-DHF-antiviral drug interactive was built with 298 nodes and 370 edges from 237 interacting genes (Supplementary Figure 2a). based on Kegg pathway genes enrichment analysis reactive ligand receptor interaction (P=3.22E-45) i.e., collection of genes associated with intracellular and extracellular signaling pathways on the plasma membrane and MAPK pathways (P=9.57E-45) that relay, amplify and integrate signals from a diverse range of stimuli and elicit an appropriate physiological response including cellular proliferation, differentiation, development, inflammatory responses and apoptosis in mammalian cells enriched the most upon antiviral drug administration (Supplementary Figure 2b). According to reactive pathway analysis indicate Phase to plateau phase (P=2.85E-34) that sustains cardiac action potential muscle contraction and transmission across chemical synapses (neurotransmitters) (P=6.03E-34) pathway genes enriches and adult (P=2.52E-69), immune system (P=1.77E-49) tissue types expressed more on antiviral drug administration on HDF according to our enrichment analysis [25,26].
Host-viral-anti-dengue hemorrhagic fever drugs target interactive
A host-DHF interactive anti-dengue hemorrhagic fever drugs interactive was built with 558 nodes and 861 edges from 419 interacting genes (Supplementary Figure 3a). Retroactive ligand receptor interaction (P=3.37E-75), cAMP signaling pathway (P=2.29E-54) also known as the adenylyl cyclase pathway, is a G protein coupled receptor-triggered signaling cascade used in cell communication are the most enriched gene pathway respectively according to Kegg pathway analysis (Supplementary Figure 3b). Amine ligand binding receptors (1.76E-47) act as neurotransmitters in humans, signal transduction (P=1.40E-46) involves the binding of extracellular signaling molecules and ligands to receptors located on the cell surface are highly enriched. Adult (P=1.74E-59), immune system (P=3.35E-54) are the most prominent tissue type during anti-dengue hemorrhagic fever drugs administration for COVID-19 patients.
Host-viral-antiviral-anti-dengue hemorrhagic fever drugs target interactive
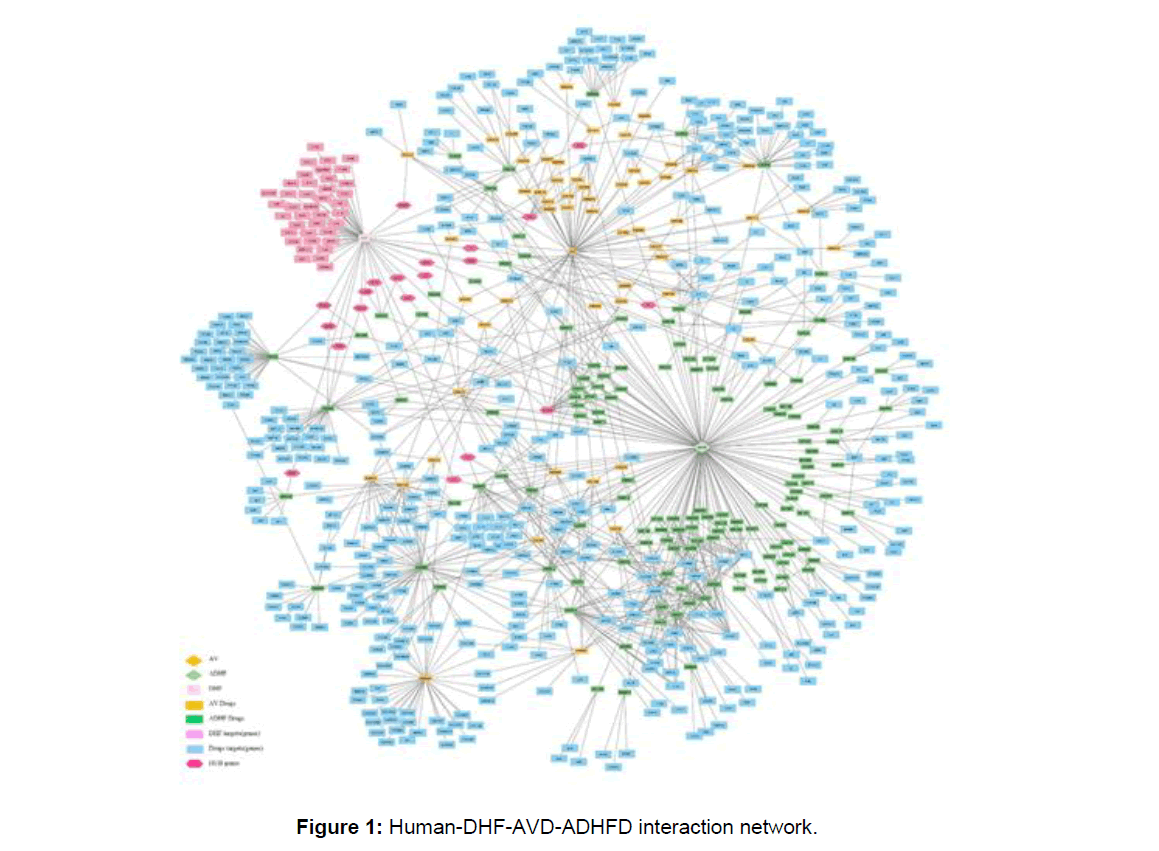
Based on all the interatomic data sets, we combine all the data sets to frame a network-based drug profiling approach to testing the robustness with which it involves network contains 717 nodes and 1175 edges from 487 interacting genes (Figure 1).
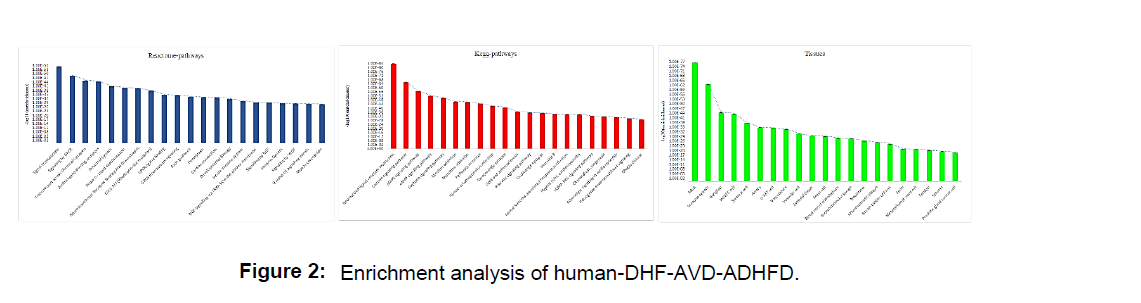
Gene functional enrichment analysis of the Kegg pathway reveals that gene sets involved in neurotransmitters pathways (P=1.13E-84) and calcium signaling pathway (P=1.78E-66) are highly enriched as similar as in previous drug related host virus interactive in human’s it also provides a stable outcome when a combined drug administration (antiviral, AHDFD) is
employed during systemic DHF patients (Figure 2). The majority of a gene set is enriched in adult (P=1.53E-77), immune system (P=3.07E-63) tissues. Genes related to signal transduction (P=6.76E-56), signaling by GPCR (P=1.96E-49) are prevalent reactive pathways enriched in DHF patients with combined drug medication.
Network based drug repurposing based on hub gene analysis
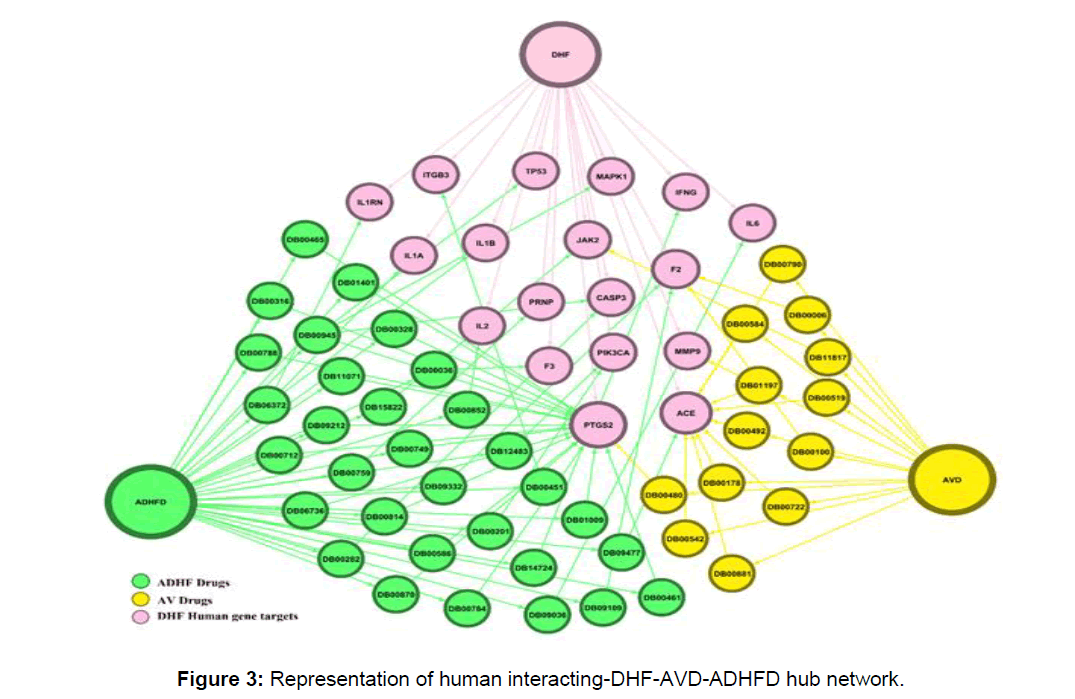
We predicted a hub gene module containing 20 interacting genes (66 nodes and 113 edges) from the above interactive of the host-virus-drugs systems framework (Figure 3).
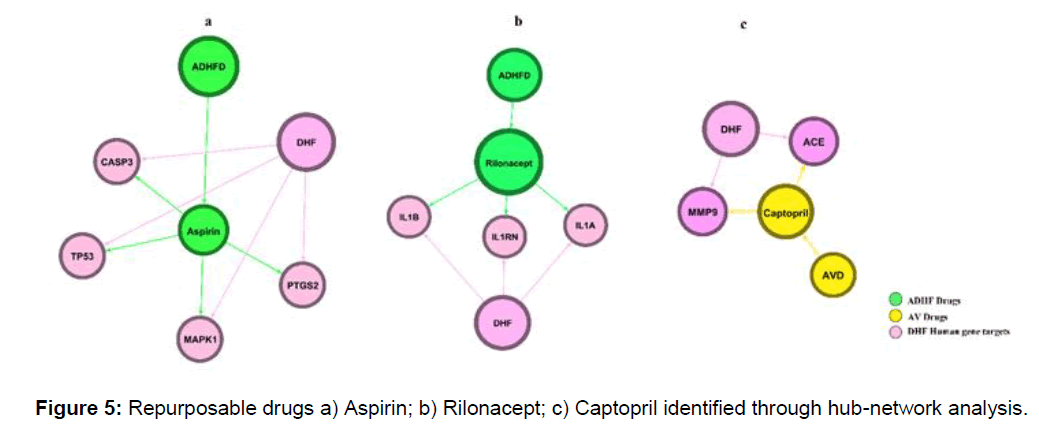
A total of 45 drugs are repurposed from the hub gene module in which 13 antiviral drugs and 32 anti-dengue hemorrhagic fever drugs. From the hub gene-drug association network we find out that 3 major drugs bind efficiently with DHF targeting human genes, aspirin, captopril, and rilonacept is efficient FDA approved drugs that can be used in the treatment of DHF (Figure 4).
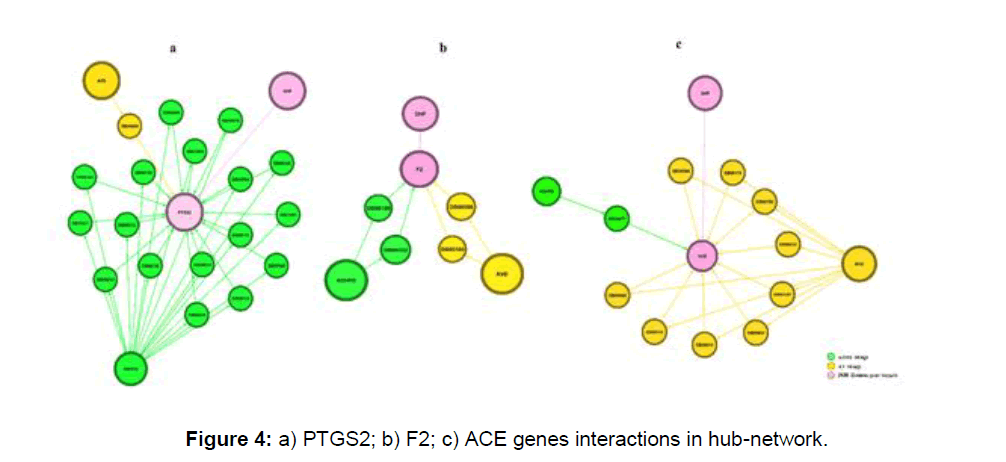
We identified 18 PTGS2 genes, 10 ACE, 4 F2, targeting drugs in hub genes in the network. Interestingly 18 out of 17 PTGS2 targeting drugs are ADHFD and 10 out 9 ACE targeting genes are antiviral drugs in property, moreover, F2 targeting drugs are equal numbers i.e., 2 AVD and 2 ADHFD (Figure 5).
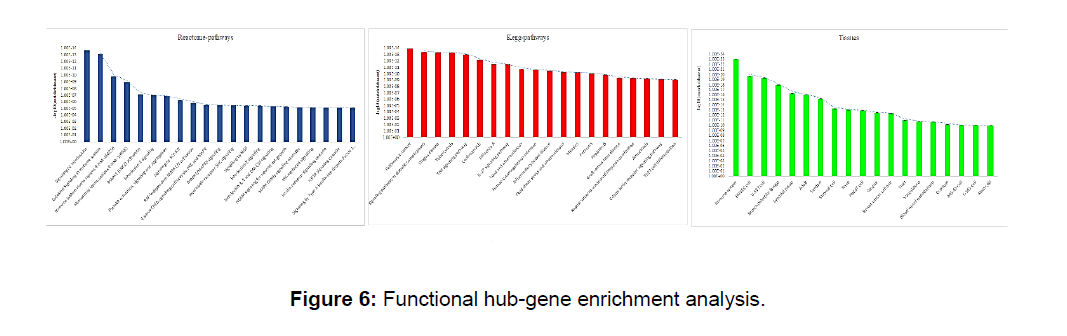
Gene enrichment analysis shows that the hub gene module is highly enriched in tissues associated with the immune system (P=7.29E-24), HUVEC cell (P=1.83E-20)this group of tissues act as an anticoagulant barrierbetween the vessel wall and blood, Kegg analysis shows that genes associated with cancer (P=1.13E-14), AGE-RAGE signaling pathway in diabetic complications (P=3.52E-14) which indicate that DHF patients with diabetes and cancer are risk of higher pathogenicity. Reactive pathway gene enrichment gives the evidence of immune systems associated pathways i.e., signaling by Interleukins (2.04E-14), cytokine signaling in immune system (7.12E-14) enrich the most (Figure 6).
Discussion
Functional enrichment analysis of drugs based on hub gene prediction
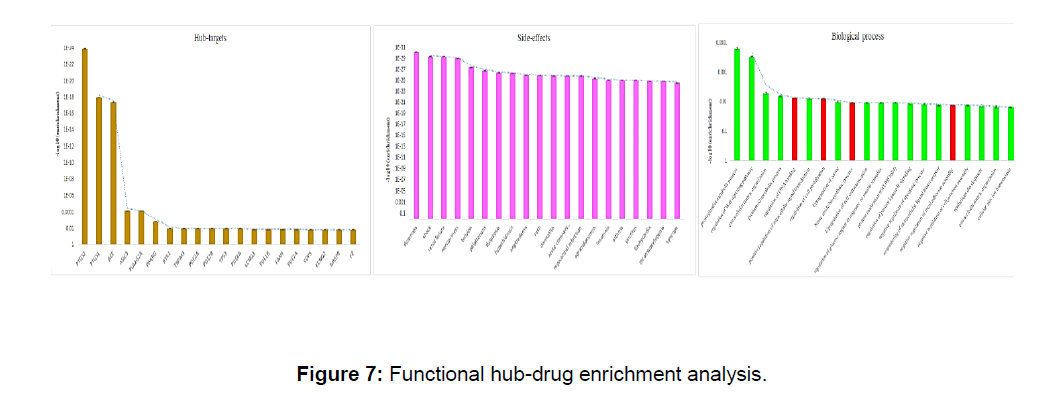
A total of 45 repurposable drugs were enriched for hub drug mechanism, gene expression of specific systems, and side effects (Figure 7). It shows that flurbiprofen, mefenamic acid, acetylsalicylic acid, indomethacin, naproxen, ketoprofen, acetaminophen, ketorolac, aceclofenac, lenalidomide, diclofenac, suprofen, loxoprofen, and nabumetone are PTGS2 targeting the hub gene module dyspnoea, shock, renal failure, nervousness, tension are prominent side effects of these drugs. Prostaglandin metabolic process (P=0.000162675), regulation of wnt signaling pathway (P=0.000307855) are prominent upregulated gene expression pathways during the above drug administration.
Conclusion
We systematically study the association of the dengue viral interaction with the human genome through network based association analysis. It is hypothesized that a host protein that functionally associates with this virus is localized in the corresponding sub-network within the comprehensive human interactive network. The host dependency factors mediating virus infection and effective molecular targets should be identified for developing broad spectrum antiviral drugs and ADHFD for DHF. In our network based analysis, we identified 45 repurposable drug candidates against DHF targeting the human gene in which 13 antiviral and 32 ADFHD that targeting 20 human genes; the most prevalent side effect identified in repurposed drug enrichment was dyspnoea and shock. PTGS2, F2, and ACE genes are the highly targeted repurposed drugs, and it already reported the pathogenicity of PTGS2/COX-2 gene pathways in the progression of DHF. Most importantly PTGS2 gene has a direct relationship with severe dengue happens when your blood vessels become damaged and leaky. And the number of clot forming cells (platelets) in your bloodstream drops. This can lead to shock, internal bleeding, organ failure and even death inhibiting this helps further prevent heart disease and better management of dengue hemorrhagic fever. For that, we sort out several effective targeting drugs based on our repurposing approach they are aceclofenac, acetaminophen, aspirin, choline magnesium trisalicylate, diclofenac, etodolac, epinephrine, indomethacin, ketoprofen, ketorolac, loxoprofen, mefenamic acid, meloxicam, nabumetone, naproxen, phenyl salicylate, suprofen, lenalidomide, enrichment analysis evidently emphasis most of the pathways of the immune system are enriched the most and adult and immune system associated tissues are associated with viral and drug response enriched throughout the study. This network based analysis hypothesized that 3 drugs have repurposable properties are aspirin, captopril, rilonacept further studies are needed to prove its efficiencies in DHF patient treatment.
Declaration of Conflicting Interests
Authors have no Conflicting Interests.
Funding Acknowledgement
Acknowledgement to “Ministry of Higher Education Malaysia for Fundamental Research Grant Scheme with Project Code: FRGS/1/2020/SKK0/USM/03/16
References
- Heilman JM, De Wolff J, Beards GM, Basden BJ (2014) Dengue fever: a Wikipedia clinical review. Open Med 8:e105–e115
[Googlescholar] [Indexed]
- Simmons CP, Farrar JJ, Nguyen VV, Wills B (2012) Dengue. N Engl J Med 366:1423-1432
[Crossref] [Googlescholar] [Indexed]
- Din M, Asghar M, Ali M (2021) COVID-19 and dengue co-epidemics: A double trouble for overburdened health systems in developing countries. J Med Virol 93:601–602
[Crossref] [Googlescholar] [Indexed]
- Sutherst RW (2004) Global change and human vulnerability to vector-borne diseases. Clin Microbiol Rev 17:136–173
[Crossref] [Googlescholar] [Indexed]
- Zhou Y, Hou Y, Shen J, Huang Y, Martin W (2020) Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov 6:14
[Crossref] [Googlescholar] [Indexed]
- Mohammadi E, Benfeitas R, Turkez H, Boren J, Nielsen J, et al. (2020) Applications of Genome-Wide Screening and Systems Biology Approaches in Drug Repositioning. Cancers (Basel) 1:2694
[Crossref] [Googlescholar] [Indexed]
- Roessler HI, Knoers NVAM, van Haelst MM, van Haaften G (2021) Drug Repurposing for Rare Diseases. Trends Pharmacol Sci 42:255-267
[Crossref] [Googlescholar] [Indexed]
- Mairiang D, Zhang H, Sodja A, Murali T, Suriyaphol P, et al. (2013) Identification of new protein interactions between dengue fever virus and its hosts, human, and mosquito. PloS one 8:e53535
[Crossref] [Googlescholar] [Indexed]
- Dey L, Mukhopadhyay A (2017) DenvInt: A database of protein-protein interactions between dengue virus and its hosts. PLoS Negl Trop Dis 11:e0005879
[Crossref] [Googlescholar] [Indexed]
- Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, et al. (2016) The Gene Cards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics. 54:1.30.1-1.30.33
[Crossref] [Googlescholar] [Indexed]
- Ramos P, Arge L, Lima N, Fukutani KF, De Queiroz A (2019) Leveraging User-Friendly Network Approaches to Extract Knowledge From High-Throughput Omics Datasets. Front Genet 10:1120
[Crossref] [Googlescholar] [Indexed]
- Zhou Y, Zhang YT, Lian XC, Zhu F, Qiu YQ, et al. (2022) Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res 50:1398-1407
[Crossref] [Googlescholar] [Indexed]
- Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, et al. (2006) Drugbank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res 34:D668–D672
[Crossref] [Googlescholar] [Indexed]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
[Crossref] [Googlescholar] [Indexed]
- Huang LJ, Law JN, Murali TM (2018) Automating the PathLinker app for Cytoscape. F1000Research 7:727
[Crossref] [Googlescholar] [Indexed]
- Groshek J, De Mees V, Eschmann R (2020) Modeling influence and community in social media data using the digital methods initiative-twitter capture and analysis toolkit (DMI-TCAT) and Gephi. MethodsX 7:101164
[Crossref] [Googlescholar] [Indexed]
- Muetze T, Goenawan IH, Wiencko HL, Bernal-Llinares M, Bryan K, et al. (2016) Contextual Hub Analysis Tool (CHAT): A Cytoscape app for identifying contextually relevant hubs in biological networks. F1000Res 5:1745
[Crossref] [Googlescholar] [Indexed]
- Freeman LC (1977) A set of measures of centrality based on betweenness. Sociometry 40:35-41
[Crossref] [Googlescholar] [Indexed]
- Brandes Ulrik (2001) "A faster algorithm for betweenness centrality." J Math Sociol 25:163-177
[Crossref] [Googlescholar] [Indexed]
- Bavelas A (1950) Communication patterns in task‐oriented groups. J Acoust Soc Am 22:725-730
[Googlescholar] [Indexed]
- Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, et al. (2021) Gene Set Knowledge Discovery with Enrichr. Curr Protoc 1:e90
[Crossref] [Googlescholar] [Indexed]
- Kay AM, Simpson CL, Stewart JA, Jr (2016) The Role of AGE/RAGE Signaling in Diabetes-Mediated Vascular Calcification. J Diabetes Res 2016:6809703
[Crossref] [Googlescholar] [Indexed]
- Park DS, Fishman GI (2011) The cardiac conduction system. Circulation 123:904-915
[Crossref] [Googlescholar] [Indexed]
- Grant AO (2009) Cardiac ion channels. Circ Arrhythm Electrophysiol 2:185-94
[Crossref] [Googlescholar] [Indexed]
- Lin CK, Tseng CK, Wu YH, Liaw CC, Lin CY, et al. (2017) Cyclooxygenase-2 facilitates dengue virus replication and serves as a potential target for developing antiviral agents. Sci Rep Mar 7:44701
[Crossref] [Googlescholar] [Indexed]
- Ruan CH, So SP, Ruan KH (2011) Inducible COX-2 dominates over COX-1 in prostacyclin biosynthesis: mechanisms of COX-2 inhibitor risk to heart disease. Life Sci 88:24-30
[Crossref] [Googlescholar] [Indexed]

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences