Identification and Sequencing of a cDNA Clone Encoding Cathelicidin-like Antimicrobial Peptide from Chicken Heart Tissues
Sundaramoorthy M,Saravanan T.S, Shivashanmugam P, Chellapandi P
Sundaramoorthy M1,Saravanan T.S2,Shivashanmugam P2,Chellapandi P3,*
1P.G. & Research Department of Zoology,AVC College,Mannampandal - 609 305,Tamil Nadu,India
2P.G. & Research Department of Zoology,Jamal Mohamed College,Tiruchirappalli-620 020,Tamil Nadu,India
3Department of Bioinformatics,Schoolof Excellence in Life Sciences,Bharathidasan University,Tiruchirappalli-620024,Tamil Nadu,India
- Corresponding Author:
- Chellapandi P
Department of Bioinformatics
Schoolof Excellence in Life Sciences
Bharathidasan University,Tiruchirappalli-620024,Tamil Nadu,India
Tel: +95- 431-2407071
Fax: +95-431-2407045
E-mail: pchellapandi@gmail.com
Abstract
In the present scenario the antimicrobial peptides of animal sources are being considered as suitable alternatives to the classical antibiotics for preventing pathogenic microorganisms without allowing them to develop resistance. Cathelicidins are one of the major classes of antimicrobial peptides widely distributed in mammalian neutrophils and epithelial cells. They kill wide-range of bacteria (both gram positive and gram negative), viruses, fungi and protozoa. In the present work, a 215 bp cDNA transcript was identified from the heart tissues of white leghorn chicken (Gallus gallus) by reverse transcription. It was amplified by RT-PCR and partially sequenced to obtain a 175 bp sequence. The sequence homology study revealed that this gene encodes a cathelicidin-like pepttide. The deduced peptide was found to be 5.5 kDa in molecular weight with 59 amino acids in length. It showed sequence similarities with cathelicidin family proteins of mammals and birds and also phylogenetically resembled with human cathelicidin-like peptide. Thus, this study will provide an insight of using this antimicrobial peptide to treat infectious diseases of human beings.
Keywords
Antibiotics; Cathelicidins; Neutrophils; LL-37; cDNA clone; Evolution; Phylogeny.
1. Introduction
Endogenous cationic peptides play an important role in innate immunity. Unlike many natural antimicrobials synthesized through specialized metabolic pathways,these peptides are expressed as precursor molecules from individual gene and converted in to active peptides by proteolytic processing. Cathelicidins are one of the major classes of antimicrobial peptides (AMPs) found in all mammalian neutrophils and epithelial cells which are evolutionarily conserved at sequence as well as structural levels. Cathelicidin peptides share a highly conserved 12kDa N-terminus named ‘cathelin’domain [1]. The C-terminals of the cathelicidin peptides express antibiotic activity after they have been cleaved from holoprotein [2].
A number of cathelicidins have been isolated and characterised from different animals. About 30 different cathelicidins like Bactenecins (Bac5 and Bac7) and PR-39 (proline-rich linear peptides) and Dodecapeptides and Protegrins (cyclic peptides) with their potential antimicrobial activities have been described in mammals [3-4]. Ambika et al.,have also identified a cathelicidin cDNA (297 base pairs in length) from the Indian goat by RT-PCR. The human cathelicidin antimicrobial peptide (hCAP-18/LL-37) was the only cathelicidin peptide identified from human neutrophils [5]. The wound healing properties of hCAP-18 [6] and is in vitro antimicrobial activities of LL-37 from ocular surface epithelial cells [7] have been investigated in detail.
Apart from mammalian cathelicidins,some avian cathelicidin peptides have also been reported. Three cathelicidin genes encoding fowlicidin 1-3 were identified from the chicken genome. They showed high degree of similarity with all known mammalian cathelicidins [8-9] have also identified an expressed sequence tag (EST) clone which encoded a novel cationic antimicrobial peptide named ‘chicken liver expressed antimicrobial peptide 2 (cLEAP-2) using in silico studies. This gene was also found to be expressed in a number of chicken epithelial tissues like small intestine,lung and kidney other than liver. Four other antimicrobial peptides,termed gallinacin 1 and 2 and turkey heterophil peptides 1 and 2 have been studied from avian heterophils [10-11]. Based on our very recent reports of antibacterial property of chicken heart tissues [12],we hypothesed that the chicken heart tissues might possess any human native antimicrobial peptides like human cathelicidin (hCAP 18/ LL 37). Hence the present study was aimed to clone and sequence the cDNA encoding human cathelicidin antimicrobial peptide from chicken heart tissues using the appropriate primers (Primers for hCAP 18/ LL 37). This work also focused on the phylogenetic inference of deduced amino acid sequence of this clone with the related sequences of vertebrates.
2. Methods
2.1 Preparation of cDNA by RT-PCR and DNA sequencing
The heart of white leghorn chicken (Gallus gallus) used in the experiments was procured from a slaughter house,Tiruchirappalli. Total RNA from the tissue sample was extracted using an RNeasy kit (Qiagen). Two hundred and fifty ng of total RNA was used for reverse transcription reaction to produce cDNA (First-Strand cDNA synthesis kit; Fermentas). Reverse transcription was performed at 54oC for 60 minutes. After denaturation of reverse transcriptase at 94oC for 5 minutes,amplification of cDNA was performed by polymerase chain reactions. Two μl of the reaction product was used for PCR amplification. The PCR reactions were performed in Eppendorf Master Cycler as per the manufacturer’s instruction (Quagen hot stat taq Master Mix kit). Total PCR volume was 16μl with 2 μl of primers per assay. The sequences of the primers for hCAP-18/LL-37 such as 5’-ATCATTGCCCAG-GTCCTCAG-3’ for forward reaction and 5’-GTCCCCATACAC-CGCTTCAC-3’ 251bp for reverse reaction were synthesised according to [7] and used to clone the human cathelicidin encoded gene. The concentration of each primer (both forward and reverse) used was 3 pmol per assay. The PCR products were visualized in the gel after the capillary electrophoresis using an Alpha Imager gel documentation system (Alpha Innotec,San Leandro,CA,USA) along with the DNA ladders between 15 to 1500 base pair lengths. The identified DNA band of 215 bp was eluted after agarose gel electrophoresis. This eluted DNA sample was further sequenced using automated DNA sequencing (ABI DNA sequence,USA) and then verified.
2.2 Sequence and structural analysis
The sequence of cDNA product was annotated for coding regions,conservd domains,motifs,structure and its biological function. The cDNA sequence was translated to corresponding amino acid sequence and then subjected to domain and motif predictions using MyHits server (https://hits.isb-sib.ch/cgi-bin/PFSCAN) from motif databases. Conserved domain classification of this sequence was searched from NCBI conserved domain architecture [13]. Pairwise similarity of this sequence with some related sequences were analyzed with LALIGN tool (https://www.ch.embnet.org/software/LALIGN_form.html). A primary and secondary structural feature of translated peptide sequence was computed by ProtParam (https://expasy.org/tools/protparam.html) and SOPMA server [14] respectively. ModWeb is an automatic comparative protein modeling server (https://salilab.org/modweb),where the same sequences were uploaded to build three dimensional structures based a template using ModPipe Version SVN.r665. A quality of modeled protein structure was further validated by SAVS (Structure Analysis and Verification) server (https://nihserver.mbi.ucla.edu/SAVS/) using Prove and ProCheck algorithms. The function of modeled protein was annotated by ProFunc server [15] by uploading PDB file as input.
2.3 Phylogenetic analysis
The highly identical sequences for cathelicidin antimicrobial peptide (CAMP) were retrieved from NCBI database using BLASTn and PSI-BLAST with default parameters [16]. Sequences with more significant identities were clustered by multiple sequence alignment program in ClustalX software [17] using Smith-Waterman substitution matrix and trimmed to consensus. Neighbor-Joining (NJ) and Minimum Evolution (ME) algorithms were used to construct phylogenic trees using MEGA 4.0 [18] with 1000 bootstraps at uniform divergence rates using Jukes and Cantor evolutionary model and 0.25 gamma distribution factors.
2.4 GenBank accession
The sequence used in this study has been deposited in GenBank of NCBI with accession number FJ755847.
3. Results
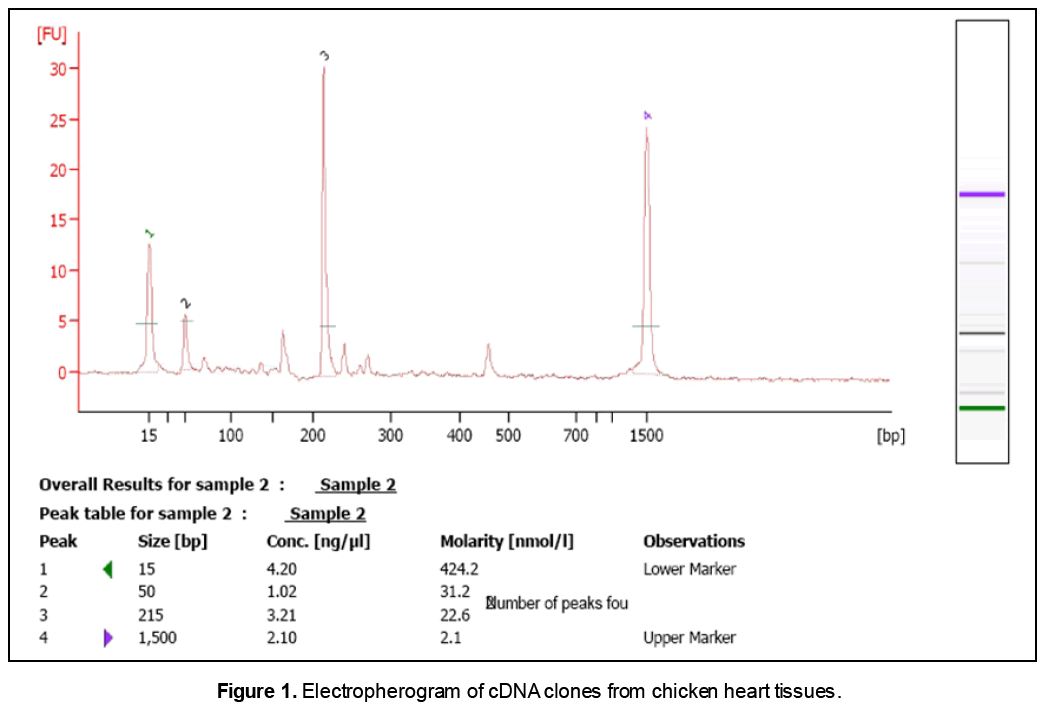
RT-PCR analysis was performed to detect the hCAP-18/LL-37 (human cathelicidin) mRNA from the chicken heart tissues. An isomer of 215 base pairs transcript was identified along with a co-amplified 50 bp cDNA clone as shown in Figure 1. Among two bands identified,the band at 215 bp transcript was distinct and dense and which was purified by agarose gel electophoresis and subjected to sequencing analysis. This was resulting into only partial sequencing that is 175bp in length instead of expected length (215bp).
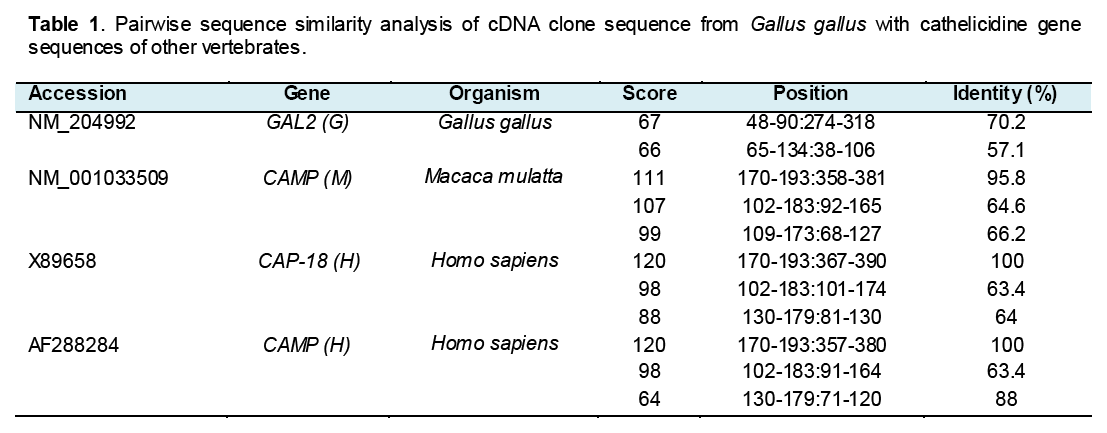
As shown in Table 1,pairwise sequence similarity analysis reported that the identified new nucleotide sequence had 100% similarities with the sequences of CAP-18 of H. sapiens at the positions of 170-193 and 367-390 where as CAMP of Macaca mulatta at the positions of 170-193 and 357-380. However,it had only 70.2% similarity with GAL2 of Gallus gallus at the positions of 48-90 and 274-318 with the score value of 67. Motif and conserved domain similarity of this sequence showed its resemblance with collagen alpha type 4 and then with hCAP of Hamo sapiens.
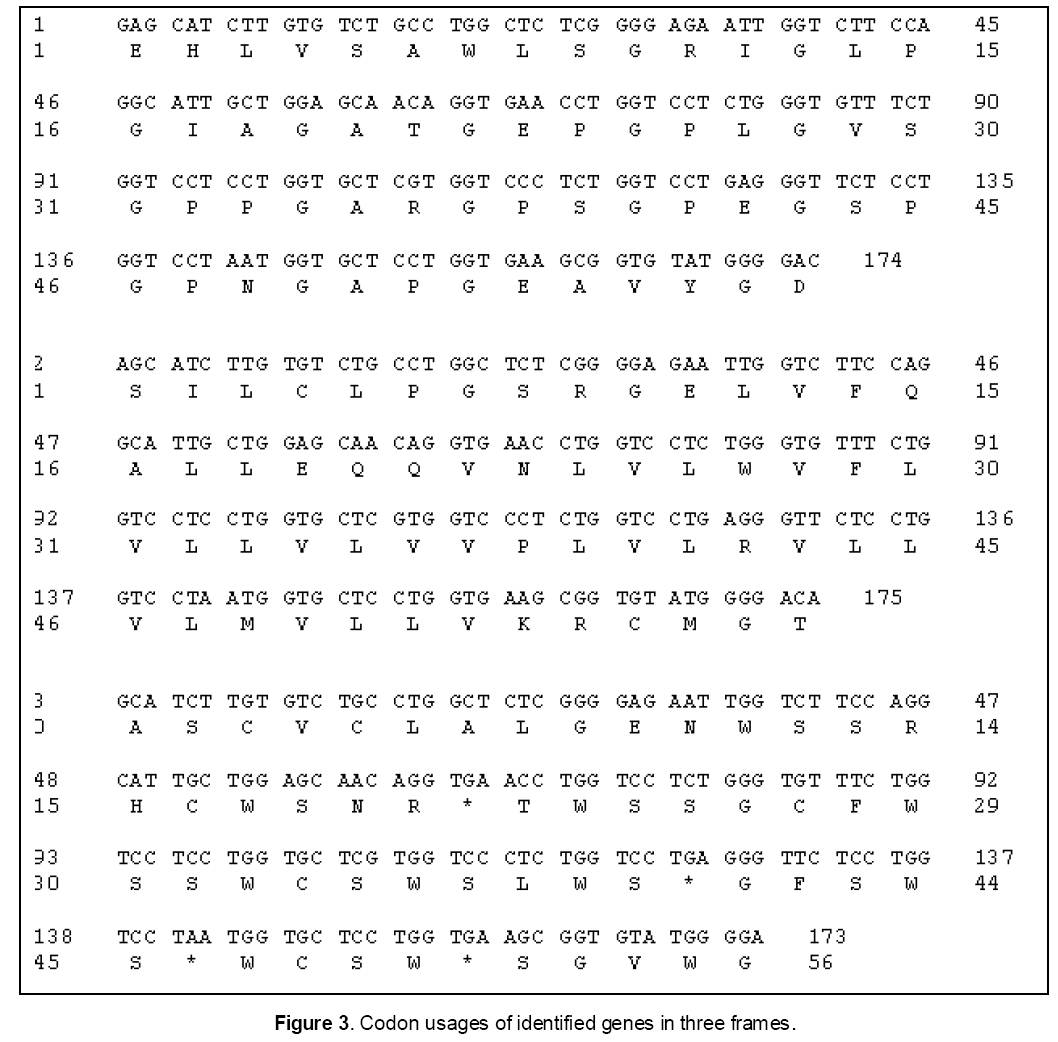
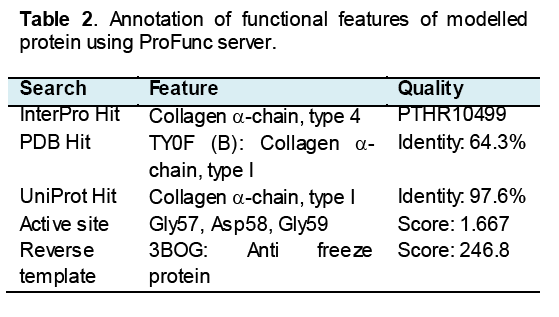
The translated peptide was found to be 5.5 kDa in molecular weight with 59 Amino acids length. It has 69.5% random coil,13.5% alpha helix,10.2% extended strand and 6.8% beta turn at secondary structure level. It was structurally aligned with the position 12-58 of template (2PNE) wherein it showed 43% identity and 0.0012 e-values. Homology modeling of this peptide showed that it has structurally homologous to collagen alpha chain type 4 (MPQS 1.34 and z-DOPE -1.17). The more conserved amino acid residues in this peptide were Gly57,Asp58 and Gly59 (Table 2). While the homology model was superimposed on hCAP-18 peptide structure,it showed the significant structural similarity and conservation of this peptide.
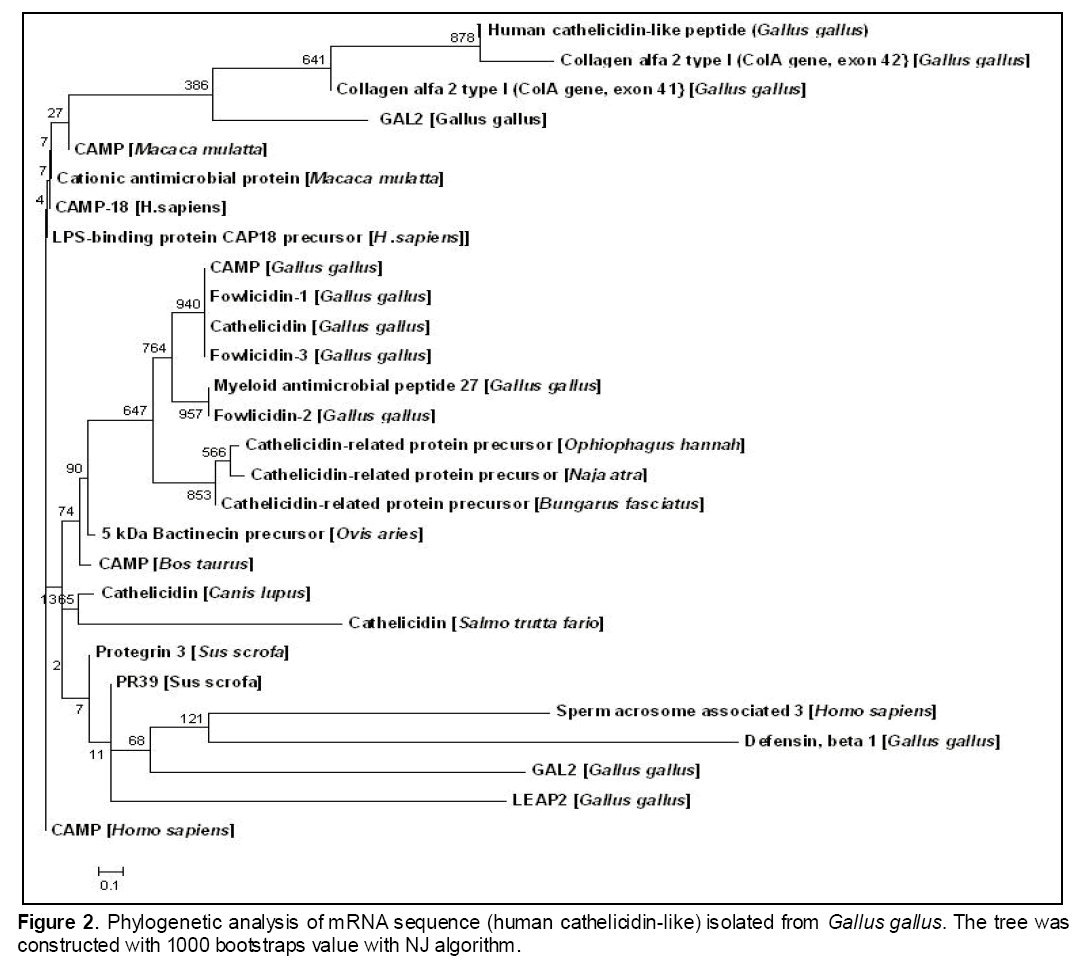
In the phylogenetic tree (Figure 2), there were 5 major clades in which human cathelicidin-like peptides from Gallus gallus was closely related with collagen alpha type 1 and 2 peptides and then clustered with gallicidin 2 of Gallus gallus and CAMP of Macaca mulatta.
This clade has phylogenetic proximity with CAP-18 and LPS-binding protein of CAP-18 of Homo sapiens,suggesting that the functional and evolutionary divergence of human cathelicidin-like peptides may be occurred. Other antimicrobial peptides,fowlicidin from Gallus gallus,bactinecin from Ovis aeries,protegrin from Sus scrofa have also shown a close phylogenetic resemblance with this peptide sequence.
4. Discussion
It is a well known fact that cathelicidins are proantibacterial proteins of neutrophils from a variety of mammalian species. The antibacterial activity of vathelicidins is confined to the C-terminal parts that differs greatly among the different cathelicidins,but share the characteristic features of being highly positively charged (cationic) and containing a high proportion of hydrophobic amino acids [19]. These features are essential for the antimicrobial activity of cathelicidins. The occurrence of such cationic amino acids and high proportion of hydrophobic regions in this peptide revealed to support the preserving antimicrobial activity against some pathogens of vertebrates.
The presence of mRNA identified for human cathelicidin (hCAP-18/LL-37) gene in the chicken heart tissues may be due to the up-regulation of this gene when the respective antibacterial peptide is depleted by utilization during infections. The up-regulation of cLEAP-2 gene in small intestine and liver of chicken after Salmonella infection has also been studied [9]. Further they have reported that birds are universally recognized as a major reservoir of human enteropathogens but are themselves often asymptomatic. Though both the nucleotide and deduced amino acid sequences of the identified cDNA clone were primarily matched with collagen type proteins,they have also significant similarities with different cathelicidin peptides including human cathelicidin. The molecular weight of the translated peptide (5.5 kDa) was approximately equal to the molecular weight of 4 to 5 kDa LL-37 functional region of human cathelicidin [20]. In addition,it was also similar to the molecular weight (5-7 kDa) of cathelicidin peptides such as Bactenecin 5 and Bactenecin 7 [21]. Hence,this cDNA clone coding peptide obtained from chicken heart tissues has suggested as a functional region of cathelicidin-like peptide (Figure 3 ). Moreover,it was also phylogenetically resembled with different cathelicidin family proteins of mammals and birds,revealing that molecular function of this peptide could be acquired from a common ancestor of birds by slow evolutionary process.
5. Conclusions
Even though the identified nucleotide sequence could not be confirmed a human cathelicidin peptide,due to all the above mentioned facts,it can be considered as a cathelicidin-like gene. Overall,we concluded that this gene isolated from chicken heart may provide a new insight to develop a potential antimicrobial peptide against pathogenic microorganisms withstanding resistance to classical antibiotics after the successful completion of full-length cDNA cloning of this gene using the appropriate cloning vectors and its expression studies. This study also reveals that it is very convenient to prepare such natural antibacterial peptides economically at industrial level.
Acknowledgements
The first author duly acknowledges the Department of Bioinformatics,Bharathidasan University,Tiruchirappalli,Tamil Nadu,India,for technical support.
References
- Ritonja A.,Kopitar M.,Jerala R.,et al. (1989) Primary structure of a new cysteine proteinase inhibitor from pig leukocytes,FEBS Letter,255: 211-214.
- Zasloff M. (2002) Antimicrobial peptides of multicellular organisms. Nature,415: 389-395.
- Gennero R.,Zanetti M. (2000) Structural features and biological activities of the cathelicidin-derived antimicrobial peptides. Biopolymers,55: 31-49.
- Ambika S.,Ashish K.,Ashok K.,et al. (2008) Cloning and characterization of goat cathelicidin cDNA. Indian Journal of Veterinary Research,17: 11-12.
- Agerberth B.,Gunne H.,Odeberg J.,et al. (1995) FALL-39 A putative human peptide antibiotic,is cystein-free and expressed in bone marrow and testis. Proceedings of the National Academy of Sciences USA,92: 195-199.
- Heilborn J.D.,Nilsson M.F.,Kratz. G.,et al. (2003) The cathelicidin antimicrobial peptide LL-37 is involved in re-epithelialisation of human skin wounds and is lacking in chronic ulcer epithelium. Journal of Investigative Dermatology,120: 379-389.
- Gordan Y.J.,Huang L.C.,Romanowski E.C.,et al. (2005) Human cathelicidin (LL-37),A multifunctional peptide is expressed by ocular surface epithelia and has potent antibacterial and antiviral activity. Current Eye Research,30: 385-394.
- Xiao Y.,Dai H.,Bommineni Y.R.,et al. (2006) Structure-activity relationships of fowlicidin-1,a cathelicidin antimicrobial peptide in chicken. FEBS Journal,273: 2581-2593.
- Townes C.L.,Michailidis G.,Nile C.J.,et al. (2004) Induction of cationic chicken liver expressed antimicrobial peptide 2 in response to Salmonella enterica infection. Infection and Immunity,72: 6987-6993.
- Evans E.W.,Beach G.G.,Wunderlich J.,et al. (1994) Isolation of antimicrobial peptides from avian Heterophils. Journal of Leukocyte Biology,56: 661-665.
- Brockus C.W.,Jackwood M.W.,Harmon B.G. (1998) Characterization of beta-defensin prepropeptide mRNA from chicken and turkey bone marrow. Animal Genetics,29: 283-289.
- Sundaramoorthy M.,Saravanan T.S. (2009) Antibacterial effects of goat and chicken heart crude tissue extracts against human pathogenic bacteria. Indian Journal of Experimental Biology,48: 407-414.
- Marchler-Bauer A.,Anderson J.B.,Cherukuri P.F.,et al. (2005) CDD: a Conserved Domain Database for protein classification. Nucleic Acids Research,33: 192-196.
- Geourjon C.,Deleage G. (1995) SOPMA: Significant Improvement in Protein Secondary Structure Prediction by consensus prediction from multiple alignments. Applied Biosciences,11: 681-684.
- Laskowski R.A.,Watson J.D.,Thornton J.M. (2005) ProFunc,A server for predicting protein function from 3D structure. Nucleic Acids Research,33: 89- 93.
- Altschul S.F.,Madden T.L.,Schaffer. A.A.,et al. (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search program. Nucleic Acids Research,25: 3389-3402.
- Thompson J.D.,Gibson T.J.,Plewniak F.,et al. (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research,24: 4876-4882.
- Tamura K.,Dudley J.,Nei M.,et al. (2007) MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution,24: 1596-1599.
- Zanetti M.,Del Sal G.,Storici P.,et al. (1993) The cDNA sequence of the neutrophil antibiotic Bac5 predicts a pro-sequence homologous to a cysteine proteinase inhibitor,that is common to other neutrophil antibiotics. Journal of Biological Chemistry,1268: 522-526.
- Sorensen O.E. (2005) The human cathelicidin hCAP-18. Danish Medical Bulletin,52: 1-10.
- Gennaro R.,Skerlavaj B.,Romeo D. (1989) Purification,composition and activity of two actenecins,antibacterial peptides of bovine neutrophils. Infection and Immunity,57: 3142-3146.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences