Genomic Analysis of Some Culex Mosquitoes (Culicidae: Diptera)
Rashmi Kohli, S. Chaudhry, Garry Bedi
1Mosquito Cytogenetics Unit,Department of Zoology,Panjab University,Chandigarh 160014,India.
2Institute of Microbial Technology,Chandigarh,160036,India
Abstract
The main objective of this study included sequence characterization of nuclear rDNA internal transcribed spacers ITS 2 and mitochondrial DNA CO II gene and 16 S gene fragment as potential molecular markers for studying genetic relatedness and phylogenetic kinship among five important species of genus Culex. After the extraction of genomic DNA from a single female mosquitoe followed by its amplification ITS 2 was found to be GC rich where as CO II and 16 S were AT rich. The incidence of indels was found to be maximum in ITS 2 and minimum in 16 S gene fragment. According to Maximum Parsimony based trees with Threshold (considering the trasnsversions) showed that Anopheles stephensi and Culex quinquefasciatus were supported by 100% bootstrap value in ITS 2 and 16 S gene fragment. But 100% bootstrap value was present between Anopheles stephensi and Culex bitaeniorhynchus in CO II gene. Beyond this all other species got bifurcated into two clades: one clade consisted of Culex tritaeniorhynchus + Culex mimeticus with bootstrap value of 62.8% while other clade consisted of Culex bitaeniorhynchus + Culex vishnui with bootstrap value of 19.5%.
Keywords
Culex; phylogeny; ITS 2; 16 S; CO II.
1. Introduction
In mosquitoes the taxonomic recognition of field collected adult specimens is often difficult because they frequently lack salient characters of scale ornamentation and the females of certain species are remarkably similar which is particularly true of the species belonging to genus Culex. Despite these limitations,the chromosome method of identification still remains an integral part for much of the contemporary work in mosquitoes systematics and population genetics. The motive behind these efforts was based on the problems of species discrimination among the species complexes in which identification of cryptic species in various taxa were problematic and frequently unsatisfactory as some of the sibling species had indistinct or poorly defined morphotaxonomic characters but very clear differences not only in their polytene chromosomal banding pattern but also in the isozymes [1],allozymes [2] and amino acid profiles.
In the recent years molecular systematics of insects has undergone remarkable growth. Polytene chromosome investigations were among the first to provide researchers with tangible genetic markers that could be used to differentiate species and their different chromosomal forms [3,4]. Besides the RAPD-PCR technique used now a days many molecular markers have been detected to understand genetic compatibility among the species [5-7]. These markers range from those located in nuclear and mitochondrial DNA.
As compared to the mosquito species belonging to genus Anopheles very little progress has been made in accumulating the genomic information on different species of the genus Culex and Aedes which also include the species of equal epidemiological significance. Keeping this in view,species specific PCR primers for genus Culex have also been designed on the basis of interspecies nucleic acid sequence variations in the first and second internal transcribed spacers (ITS 1 and ITS 2) of nuclear rDNA gene array to differentiate between subspecies of Culex pipiens complex. Ribosomal DNA spacers are also a class of important genetic markers for developing PCR based species diagnostic [8,9]. Preceding the 18S gene is the external transcribed spacer (ETS) and surrounding the 5.8 S rDNA are the internal transcribed spacers 1 and 2 (ITS 1,ITS 2). Studies conducted so far have revealed that this multigene family evolves collectively within species,a mechanism that tends to produce homologous nucleotide sequences within species while producing genetic differences among them [10]. The coding sequences are highly conserved even between distantly related species,while non-coding sequences rapidly drifts apart even among the closely related species. Thus using suitable primers variable regions of rDNA sequences can be amplified without the prior sequence information. As such the rDNA gene cluster has become an increasingly popular tool in molecular entomology and a means of differentiating cryptic anopheline and nonanopheline species of mosquitoes [11].
Rapid evolution of rDNA within species has resulted in the use of faster evolving spacers,not only for the reconstruction of mosquito phylogenies but also as diagnostic markers for differentiating cryptic species of anophelines [8,12,13,14,15]. ITS 2 sequences are relatively short,less than 1 kilobase pairs (kbp),making the amplification of intervening ITS 2 using specific matching primers from highly conserved domains of flanking coding regions (5.8 S and 28 S respectively) relatively simple. Second,the level of intraspecific variations in them is lower than the interspecific variations.
Apart from the nuclear DNA,the mitochondrial DNA has its own unique properties for which it has been found quite a useful alternative in those genomic studies where there are differences and homologies in the genetic make up of a species. The small size of mitochondrial genome,its single copy number,lack of introns and maternal inheritance are some of its features which are preferred for DNA diagnostics.
2. Materials and Methods
Inspired by work done in molecular cytogenetics of mosquitoes,the present topic of research on some Culex mosquitoes (Diptera: Culicidae) was undertaken to carry out sequence based phylogenetic inferences of five epidemiologically important species of genus Culex viz: Culex quinquefasciatus,Culex vishnui,Culex bitaeniorhynchus,Culex tritaeniorhynchus and Culex mimeticus.
The objective of the study included sequence characterization of ITS 2,CO II and 16 S gene sequence as potential molecular markers. Anopheles stephensi was used as an outgroup so as to validate the results. The field collection of these species was carried out from a village Nadasahib (Panchkula,Haryana),20 Kms South- East of Chandigarh,Hamirpur (Himachal Pradesh),Patiala,(Punjab) and Sector 25 of Chandigarh.
Specimens belonging to different species were collected at random and brought to the laboratory in a small field cage.
3. Results
3.1 Sequence Analysis of ITS 2
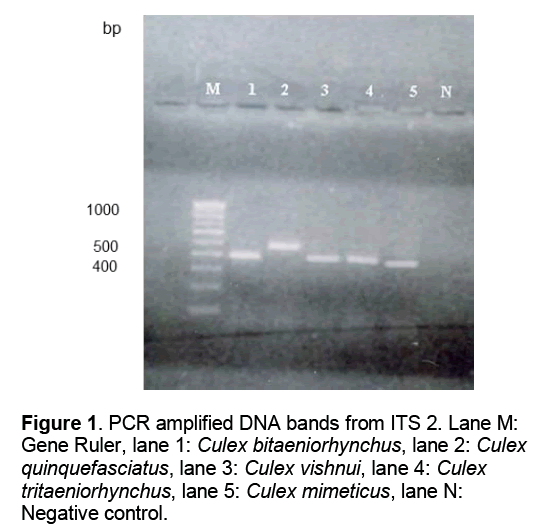
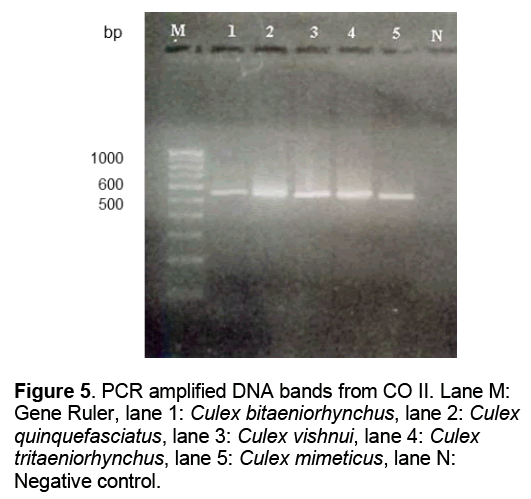
PCR amplification of genomic DNA with ITS 2 primers in each of the samples across the five species yielded fragments of 390- 477 base pairs. In addition to this Anopheles stephensi was taken as an outgroup to establish the relationship between anophelines and non-anophelines. In the gel lanes 1-5 were loaded with amplified PCR products of Culex bitaeniorhynchus,Culex quinquefasciatus,Culex vishnui,Culex tritaeniorhynchus and Culex mimeticus respectively. Lane M was loaded with 100 bp gene ruler while lane N carried a negative control (Figure 1).
The detailed nucleotide sequence analysis of ITS 2 gene sequence revealed marginally higher percentage 51.8% of G+C content in each of the species as compared to 48.2% of A+T content in each species hence the ITS 2 gene was found to be GC rich.
As per multiple sequence alignment it was found that ITS 2 sequence showed indels at 221 positions in the sequence and the transition to transversion ratio (ts/tv) ranged from 0.41 to 0.76,while Cx. tritaeniorhynchus had the minimum ts/tv value of 0.4 and Cx. mimeticus was found to have the maximum ts/tv value of 0.76. In fact,transitions are the loci where purines get replaced by purines while transversions are the sites where purines get replaced by pyrimidines and vice-versa. According to the rate of substitution,the transversions were found to be more than transitions. The most frequent transversions were of the A-T type.
3.2 Phylogenetic analysis on the basis of sequence homologies of ITS 2 nuclear gene
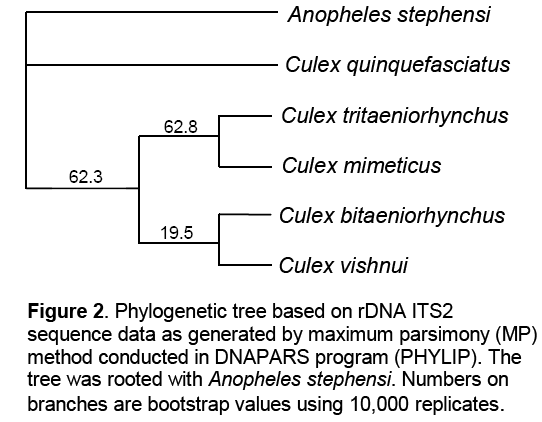
According to the Maximum Parsimony (MP) based tree with Threshold (considering the transversions) showed that An. stephensi and Cx. quinqefasciatus were supported by 100% bootstrap value beyond which all other species got bifurcated into two clades: one clade consisting of Cx. tritaeniorhynchus + Cx. mimeticus with a bootstrap value of 62.8% while other clade consisted of Cx. bitaeniorhynchus + Cx. vishnui with a bootstrap value of 19.5%. In order to assess the branch support,100,000 bootstrap [16] replications were completed. This is a method to estimate confidence levels of inferred relationships (Figure 2 ).
3.3 Sequence Analysis of CO II gene
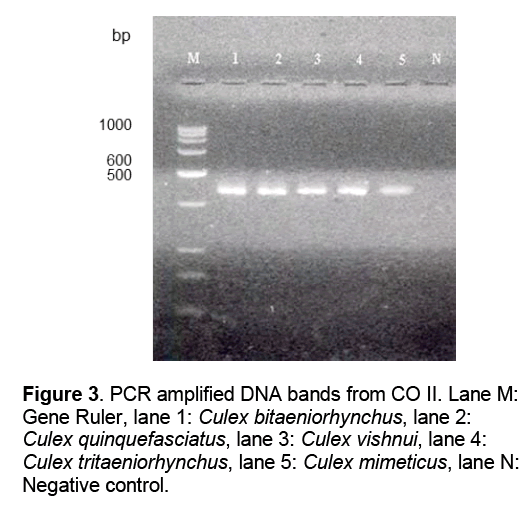
Bands of approximately 540 base pairs were generated (Figure 3).
The detailed nucleotide sequence analysis of CO II mitochondrial gene revealed significantly higher percentage of A+T (75.2%) base composition as compared to G+C (24.8%) therefore it could be labeled as an AT rich sequence.
It was seen that as many as 198 positions showed indels. The transition to transversion ratio (ts/tv) ranged from 0.10 to 0.81 with Cx. mimeticus with the least ts/tv value of 0.10 while An. stephensi with the highest ts/tv value of 0.81.
Moreover,the base substitution ratio showed more transitions as compared to transversions. The most frequent transversions were of T-C type.
3.4 Phylogenetic analysis on the basis of sequence homologies of CO II mitochondrial gene
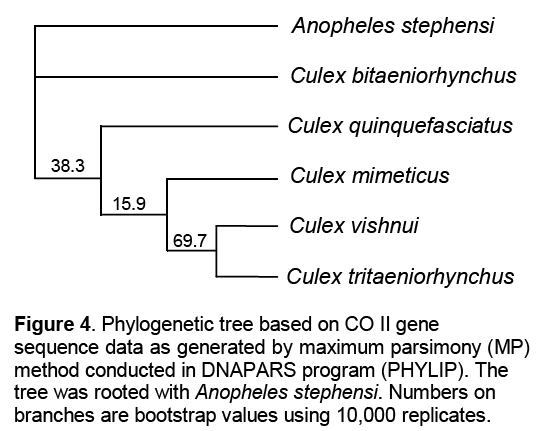
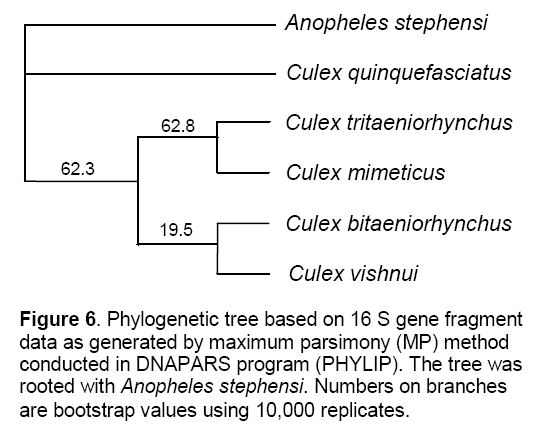
The phylogenetic tree constructed using Maximum Parsimony Method showed 100% bootstrap value between An. stephensi and Cx. bitaeniorhynchus,whereas Cx. quinquefasciatus and Cx. mimeticus were supported by weak bootsrtap values of 38.3% and 15.9% respectively. Cx. vishnui and Cx. tritaeniorhynchus were bifurcated into a separate clade with a bootstrap value of 69.7% (Figure 4 ).
sequence data as generated by maximum parsimony (MP) method conducted in DNAPARS program (PHYLIP). The tree was rooted with Anopheles stephensi. Numbers on branches are bootstrap values using 10,000 replicates.
3.5 Sequence Analysis of 16 S gene
The PCR amplification of 16 S mitochondrial gene yielded 550 base pairs long fragments with the specific primers (Figure 5 ).
Multiple sequence alignment was carried out using Clustal W so as to edit the sequences to the same length in order to reduce the avoidable extensions of the sequences. The sequence was found to be AT rich and the indels were observed at 165 positions. The statistical analysis of the alignment data showed that the transition to transversion ratios of the bases (ts/tv) ranged from 0.10 to 0.66 with Cx. tritaeniorhynchus and Cx. quinquefasciatus having ts/tv value of 0.10 while Cx. bitaeniorhynchus having the highest ts/tv value of 0.66. Transversions were found to be more than transitions and A-T type were more frequent. The detailed nucleotide sequence analysis of 16S gene revealed considerably higher A+T content of 81.4% as compared to G+C content of only 27.6%.
3.6 Phylogenetic analysis on the basis of sequence homologies of 16S mitochondrial gene
According to Maximum Parsimony Method the bootstrap value was 100% between An. stephensi and Cx. quinquefasciatus,whereas Cx. tritaeniorhynchus have just 75% bootstrap value. Similarly,Cx. bitaeniorhynchus was also found to have absolute bootstrap value of 100% and that placed it in proximity to a clade including Cx. mimeticus and Cx. vishnui with a 99% bootstrap value (Figure 6 ).
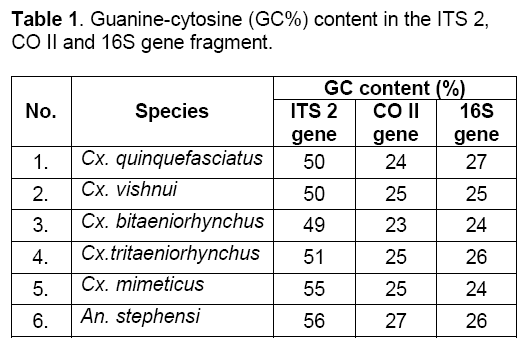
Calculations from multiple sequence alignment showed minimum GC % content in Culex bitaeniorhynchus and maximum in Anopheles stephensi in all the three genes i.e. ITS 2,CO II and 16S gene fragment (Table 1).
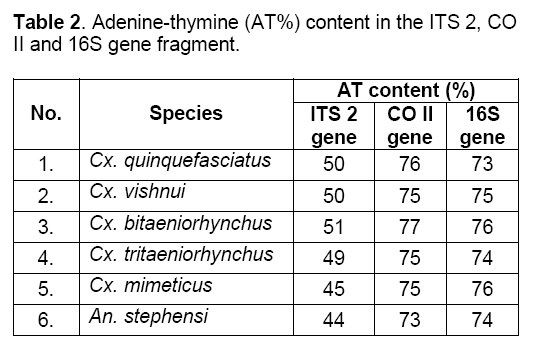
Calculations of AT % content showed minimum value in Anopheles stephensi and maximum in Culex bitaeniorhynchus (Table 2).
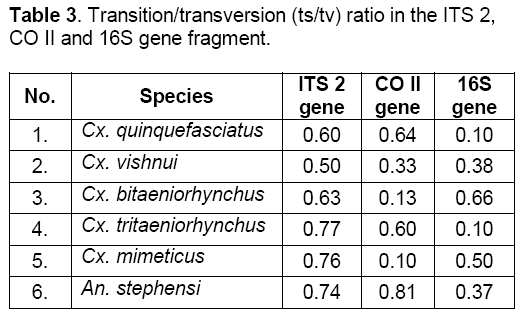
Mean of the ts/tv ratio was 0.666 in ITS 2 gene,0.435 in CO II gene and 0.351 in 16S gene fragment (Table 3). These results showed that ITS 2 gene was prone to maximum base pair replacements. Hence,being a nuclear gene it was least conserved followed by CO II and 16S gene fragment respectively. Being mitochondrial genes they were more conserved and less prone to base substitutions i.e. transitions (ts) and transversions (tv).
4. Discussions
As a result of the present research work,the following points of interests in the study of mosquito genomics and accomplishments in the molecular cytogenetics of these insects need a special mention.
It is the first ever attempt on the sequence analysis of the five species of genus Culex from the Oriental Region including the Indian subcontinent. It is for the first time that sequence variations of three genes (ITS 2,CO II and 16S rRNA) have been studied collectively. Out of the three studied genes,two genes were AT rich. In transition to transversion (ts/tv) ratios,the transversion frequencies were more than transition frequencies. As for the incidence of insertions/deletions (indels) of bases is concerned,it was found maximum in ITS 2 gene and minimum in conserved mitochondrial genes CO II and 16S rRNA. It is for the first time that in addition to interspecific comparisons,intergeneric comparisons were carried out by taking one Anopheles species as an outgroup. As many as 15 gene sequences were submitted to Genbank and accession numbers have been acquired. ITS 2 gene sequence was found to be most ideal for identification of sibling species in genus Culex.
5. Conclusions
The dendrograms clearly show that Cx. vishnui and Cx. tritaeniorhynchus fall under one clade with close proximity to Cx. quinquefasciatus and Cx. bitaeniorhynchus trailing behind,whereas Cx. mimeticus shows closeness with An. stephensi instead of either of the Culex species.
Acknowledgements
The authors are thankful to Chairperson,Department of Zoology,Panjab University,Chandigarh for providing the necessary facilities under Centre of Advance Studies Programme of U.G.C,New Delhi,India to carry out the present research work.
References
- Adak T.,Subbarao S.K.,Sharma V.P.,Rao S.R. (1994) Lactate dehydrogenase allozyme differentiation of species in the Anopheles culicifacies complex. Med. Vet. Entomol.,8(2): 137-140.
- Coosemans M.,Smits A.,Roelants P. (1998) Intraspecific isozyme polymorphism of An. gambiae in relation to environment,behaviour and malaria transmission in South-western Burkina Faso. Am. J. Trop. Med. Hyg.,58: 70-74.
- Coluzzi M.,Sabatini A.,Petrarca V.,Di Deco M.A. (1979) Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans. R. Soc. Trop. Med. Hyg.,73: 483-497.
- Toure Y.T.,Petrarca V.,Traore S.F.,Coulibaly A.,Maiga H.M. (1998) The distribution and inversion polymorphism of chromosomally recognized taxa of the Anopheles gambiae complex in Mali,West Africa. Parasitologia,40: 477-511.
- Williams J.G.K.,Kubeklik A.R.,Livak K.J.,Rafalski J.A.,Tingey S.V. (1990) DNA polymorphisms amplified by arbitrary primers are useful genetic markers. Nucleic Acids Res.,18: 6531-6535.
- Favia G.,Dimopoulos G.,Della T.A.,Toure Y.T.,Coluzzi M.,Louis C. (1994) Polymorphisms detected by random PCR distinguish between different chromosomal forms of Anopheles gambiae. PNAS,91: 10315-10319.
- Munstermann L.E.,Conn J.E. (1997) Systematics of mosquito disease vectors (Diptera : Culicidae): impact of molecular biology and cladistic analysis. Annu. Rev. Entomol.,42: 351-369.
- Cornel A.J.,Porter C.H.,Collins F.H. (1996) Polymerase chain reaction species diagnostic assay for Anopheles quadrimaculatus cryptic species (Diptera: Culicidae) based on ribosomal DNA ITS 2 sequences. J. Med. Entomol.,33(1): 109-116.
- Li C.,Wilkerson R.C. (2005) Identification of Anopheles (Nyssorhynchus) albitarsis complex species (Diptera: Culicidae) using rDNA internal transcribed spacer2-based polymerase chain reaction primes. Mem. Inst. Oswaldo Cruz.,100(5): 495-500.
- Collins F.H.,Paskewitz S.M. (1996) A review of the use of ribosomal DNA (rDNA) to differentiate among cryptic Anopheles species. Insect Mol. Biol.,5(1): 1-9.
- Collins F.H.,Mendez M.A.,Rasmussen M.O.,Mehaffey P.C.,Besansky N.J.,Finnerty,V. (1987) A ribosomal RNA gene probe differentiates member species of the Anopheles gambiae complex. Am. J. Trop. Med. Hyg.,37: 37-41.
- Wesson D.M.,Porter C.H.,Collins F.H. (1992) Sequence and secondary structure comparisons of ITS rDNA in mosquitoes (Diptera:Culicidae). Mol. Phylogen. Evol.,1: 253-269.
- Scott J.A.,Brogdon W.G.,Collins F.H. 1993. Identification of single specimen of the Anopheles gambiae complex by the polymerase chain reaction. Am. J. Trop. Med. Hyg.,49: 520-529.
- Porter C.H.,Collins F.H. (1996) Phylogeny of nearctic members of the Anopheles maculipennis species group derived from the D2 variable region of 28S ribosomal RNA. Mol. Phylogenet. Evol.,6: 178-188.
- Felsenstein J. (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution,39: 783-791.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences