Genome-wide identification of PHYTOCHELATIN and PHYTOCH_SYNTH domain-containing phytochelatin family from rice
Guo-ming Shen, Cheng Zhu, Qi-zhen Du
Guo-ming Shen1,3,Cheng Zhu2,*,Qi-zhen Du3
1State Key Laboratory of Plant Physiology & Biochemistry,College of Life Sciences,Zhejiang University,Hangzhou,310058 China
2College of Life Sciences,China Jiliang University,Hangzhou,310018 China
3School of Food Science and Biotechnology,Zhejiang Gongshang University,149 Jiaogong Road,Hangzhou,310012 China.
- Corresponding Author:
- Cheng Zhu
Tel: +86(0)571 8691 4510
Fax: +86(0)571 8691 4510
E-mail: pzhch@cjlu.edu.cn
Abstract
Phytochelatin synthases (PCS) are cytosol proteins that may play vital roles in heavy metal detoxicity in bacteria, yeasts, plants, or worms. More than 10 PCS-encoded genes from different species have been reported during the last decade, and they were played critical roles in cadmium or arsine detoxfication. In this investigation, we identified 3 putative PCS and 11 putative PCS-like genes in rice genome by the bioinformatics analysis of recently completed rice genome data, classified them into 2 subfamilies on the basis of domain sequence similarity by constructing phylogenetic trees, and localized them in rice chromosomes. Reverse transcription polymerase chain reaction (RT-PCR) was performed to demonstrate that the expression of 3 putative PCS genes (OsPCS3, OsPCS8, and OsPCS11) could not be detected in the roots, stem base, nodes, internodes or petioles, but all the remaining 11 putative rice PCS genes were found to be expressed in the leaves and panicles, OsPCS7 was induced by Hg2+ and Pb2+, while OsPCS9 was induced by Cd2+ and Zn2+ in roots, suggesting that PCS or PCS-like genes might play crucial roles in survival or in different heavy metal detoxification
Keywords
Gene family; heavy metal detoxification; phylogenetic evolution; phytochelatin synthase; Oryza sativa.
1. Introduction
Phytochelatins (PCs) are a class of post-translationally synthesized peptides that may play a vital role in heavy metal detoxification and accumulation in plants [1-3]. PCs combine heavy metals such as cadmium (Cd) or arsine (As) with high affinity,and localize themselves bound with the heavy metal ions to the cell vacuoles,thus playing a role in detoxification [1,2]. Their synthesis is mediated by phytochelatin synthase (PCS) (γ-glutamylcysteine dipeptidyltrans-peptidase,EC 2.3.2.15) using glutathione (GSH) and related thiol tripeptides as substrates,transfering a γ-Glu-Cys unit from one thiol peptide to another or to pre-existing phytochelatin molecules [4,5]. PCS is activated by heavy metals,such as Cd [4].
PCS genes firstly were cloned by three independent laboratories with different methods [6-8]. In plants,there had been reported PCS genes in Arabidopsis,wheat (Triticum aestivum L.),Thlaspi caerulescens L. J. & C. Presl and Thlaspi japonicum H.,soybean (Glycine max L. Merr.),Nostoc sp. PCC 7120 L.,Pteris vittata L.,lettuce (Lactuca sativa L.),lotus (Lotus japonicus L.),rice (Oryza sativa L.) [3,6-15] and so on. Overexpression of Arabidopsis thaliana phytochelatin synthase (AtPCS1) gene in Escherichia coli (T. Escherich) or Mesorhizobium huakuii (Chen) enhanced the accumulation of heavy metals in those bacteria [16,17],while,overspression of AtPCS1 in zebrafish (Danio rerio Hamilton-Buchanan) or tobacco (Nicotiana tobacum L.) enhanced heavy metal tolerance [18,19]. Interestingly,overexpression of AtPCS1 in Arabidopsis was paradoxically hypersensitive to cadmium [20,21]. Therefore,the functions of PCS genes and their homologues and their roles in heavy metal detoxification need to be further investigated in other plants.
The recent completion of the rice genome-sequencing project,together with its automated annotation process,enabled us for the first time to gauge the number of PCS family with a typical angiosperm [22]. In this report,we identified 3 PCS and 11 PCS-like genes in rice genome using bioinformatics methods,and analyzed their expression patterns by Reverse transcription polymerase chain reaction (RT-PCR).
2. Materials and Methods
Plant materials
Rice (Oryza sativa L. ssp japonica variety,Zhonghua 11) seeds were surface sterilized in 0.5% sodium hypochlorite for 20 min,rinsed,and germinated in the dark on moistened filter paper at 30°C for 2 d,and then transferred the germinated seeds to 96-hole plastic floatings for 4 d. The uniformly germinated seedlings were transferred to black polyethylene barrels which contain 8 L rice culture solution [23] in growth chambers (MC1000 system; Snijders) at temperature regimes of 30/24°C (day/night) and 70% humidity under a 12-h photoperiod (photo flux density of 500 μM s-1 m-2) during the growth period,the nutrient solution was replaced every five day. The roots,stembase,node,internode,petiole,leaves,and panicles were harvested and dipped in liquid nitrogen and then stored at -80°C until RNA extraction. For metal-induced gene expression experiment,9-d old seedlings were transferred to half-strength rice culture solutions containing 100 mM CdCl2,100 mM CuCl2·2H2O,100 mM ZnSO4·7H2O,100 mM HgCl2,100 mM MnCl2,100 mM Pb(NO3)2,100 mM CoCl2 or 100 mM AgNO3,respectively,for 1 d. The roots were harvested and dipped in liquid nitrogen and then stored at -80°C until RNA extraction.
cDNA synthesis and RT-PCR analysis
Total RNA was extracted from the roots,stembase,nodes,internodes,petioles,leaves and panicles in mature plants using Trizol reagent (Invitrogen,USA). cDNA synthesis was performed as described by Reale et al [24]. Reverse transcription polymerase chain reaction (RT-PCR) was conducted in a 25 μL reaction containing 20 pmole of OsTUB16 (accession number X78143) primers (5’-CGCCT CTGCCATGTTCCGTGGAA-3’ and 5’-GGCGGTAA TACGGTGATAATGTAA-3’),and gene-specific primers (see Supplemental Table 1),10 mM dNTPs,5 unit of Ex-Taq DNA polymerase (TakaRa,Japan),and 10×reaction buffer. RT-PCR was performed under optimal conditions for each gene,and the numbers of reaction cycles were 33~35. Afterward,5 μL aliquots of the reaction mixtures were separated on 1.2% (w/v) agarose gels.
Sequence analysis and construction of the phylogenetic tree
Database searching,editing,and sequence alignments described as Cai and Lytton [25],sequence analysis of a variety in molecular evolutionary genetics analysis (MEGA) version 4.0 using the ClustalW program,building phylogenetic trees also use the MEGA 4.0 software package. Calculating the sequences of different base composition,the percentage of variation sites,codon usage,the percentage of conversions and transversions,or the percentage of transversions as the methods described by Tamura [26]. Two-parameter method to calculate the genetic distance between branches,using neighbor-joining,minimum evolution and maximum parsimony method for system reconstruction as described by Saitou and Nei [27]. Phylogenetic tree of the confidence level of the branch by re-sampling method (bootstrap) 1000 repeated testing,DNA sequence variation of the conversions and transversions given the same valuation were used in this investigation.
Determination of conserved domain
Conservative functional domain analysis was carried out by using DNAMAN version 6.0 (Lynnon corpora-tion,Vaudreuil-Dorion,Canada),Hidden Markov Model (HMM) [28] and on-line version of GlobPlotTM 2.3 software package (https://globplot.embl.de/) [29].
3. Results
3.1 Identification and structure of PCS and PCS-lick genes in rice
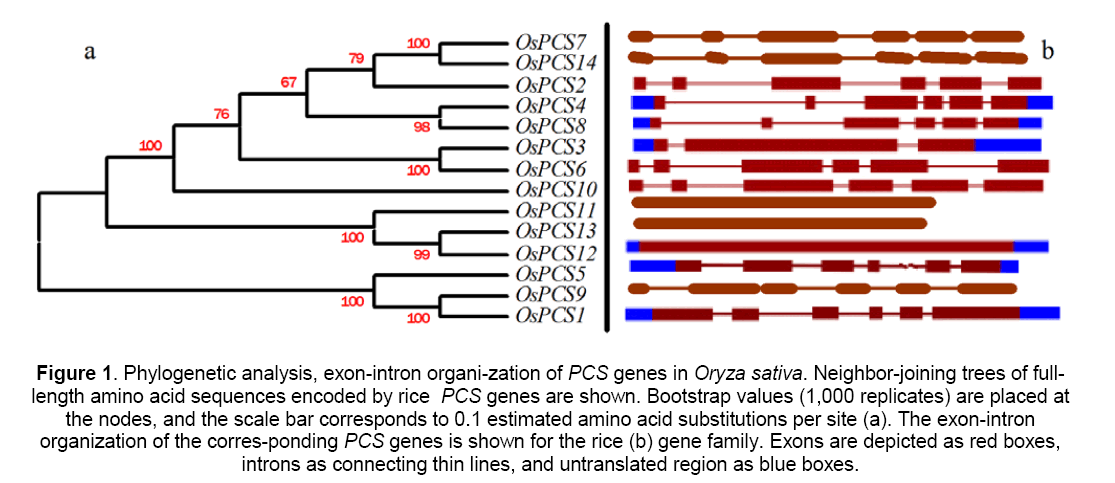
To identify the PCS family genes in rice,basic local alignment search tool (BLAST) [30] searches of the rice databases were performed using the PHYTOCHELATIN domain (pfam05023) of the Arabidopsis AtPCS1 protein as a query sequence. Three genes were identified to possibly encode PHYTOCHELATIN domain,namely,OsPCS1,5 and 9 (Table 1). The search of “gene” using “phytochelatin synthase (PCS)” as a keyword in NCBI matched 7 homologues of OsPCS 2-4,6,8,10 and 12. tBLASTn searches of the rice database were performed using the rice OsPCS2 protein as a query sequence,and other 4 genes were matched. In order to predict the genetic classification of PCS in this study,we have built phylogenetic trees based on the full-length amino acid sequences [26],analysis showed that: the predicted 14 genes can be divided into two sub-group as showed in Figure 1a. In RiceGAAS (https:// ricegaas.dna.affrc.go.jp) web site on our forecast of 14 genes in exon-intron composition analysis results showed that 10 of the 14 OsPCS genes have 6 exons,1 has 3 exons,and the other 3 have no intron (Figure1b).
Figure 1: Phylogenetic analysis, exon-intron organi-zation of PCS genes in Oryza sativa. Neighbor-joining trees of full-length amino acid sequences encoded by rice PCS genes are shown. Bootstrap values (1,000 replicates) are placed at the nodes, and the scale bar corresponds to 0.1 estimated amino acid substitutions per site (a). The exon-intron organization of the corres-ponding PCS genes is shown for the rice (b) gene family. Exons are depicted as red boxes, introns as connecting thin lines, and untranslated region as blue boxes.
We explored protein sequences for intrinsic protein disorder,domain and globularity prediction by software GlobPlotTM version 2.3. Unfortunately,PCS1,5,13 or 14 were not detected PHYTO-CHELATIN or PHYTOCHEL_SYNTH domain (see Supplemental Figure 1).
3.2 Chromosomal distribution
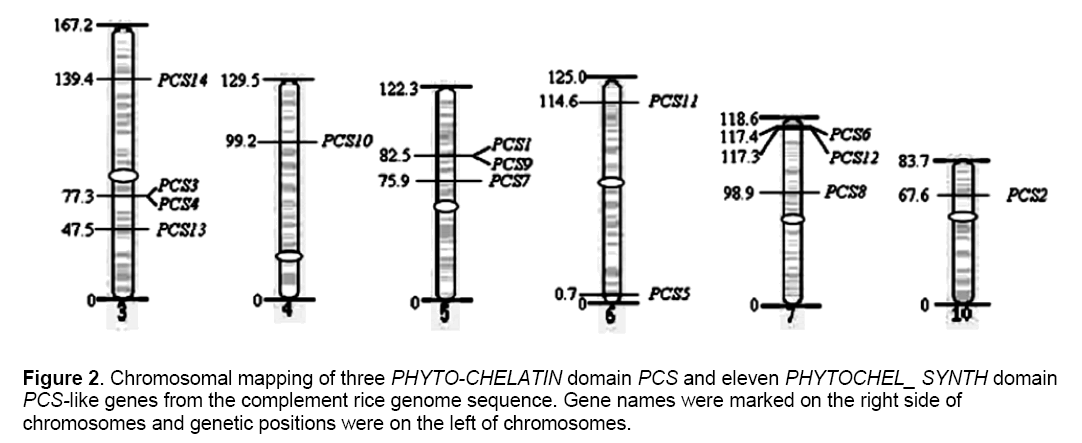
We used the data from Rice Genome Research Program (GRP) website (https:// rgp.dna.affrc.go.jp/) on the corresponding YAC or BAC cloning genetic map database to localize the map position of the predicated PCS genes [22]. Figure 2 showed that the distribution of the 14 OsPCS genes on different chromosomes in the rice genome. All of the predicted OsPCS sequences were distributed within chromosomes 3~7 and 10,and no homologues were located at other chromosomes. Although the 6 chromosomes had homologues,each differed in the number of OsPCS genes it contained chromo-somes 4 and 10 each contained only 1 OsPCS gene,chromosome 6 contained 2,chromosomes 5 and 7 each contained 3,and chromosome 3 had 4. OsPCS 3-5 and 13 oriented the of short arm of rice chromosome,and the rest located in the long arm of rice chromosome,while OsPCS 3,4,7 and 8 is located near the centromere,OsPCS 5,6 and 12 located near the end of the long arm (from the end of the genetic map distance of less than 2 centi Morgan).
3.3 Expression pattern of PCS and PCS-like genes
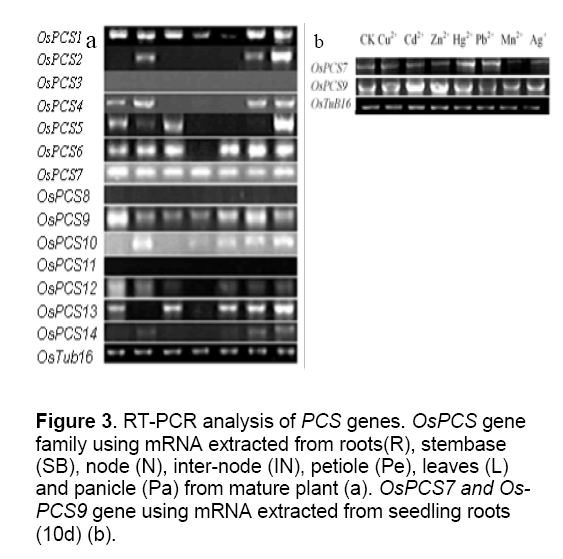
In order to verify whether the predicted genes can be truly expressed and to know their expression patterns,we have extracted RNA in different organs from the rice. Before reverse transcription,RNAs were treated with RNase-free Dnase to avoid genomic DNA contamination for the reliability of the experiment. Primers were designed based on coding open reading frame regions of PCS genes and are shown in Supplemental Table 1. The RT-PCR results show that not all predicted genes were expressed in tissues tested under normal growth conditions. OsPCS3,OsPCS8,OsPCS11 genes could not be detected even if we modified the RT-PCR conditions. The other 11 OsPCS genes expressed almost in all the leaves and panicles,and different expression patterns were observed in the roots,stem base,nodes,internodes,and petioles (Figure 3 a). To investigate metal inducing gene expression pattern,we selected OsPCS7 standing for the PCS-like subfamily and OsPCS9 standing for the PCS family. OsPCS7 was induced by Hg2+ and Pb2+,while OsPCS9 was induced by Cd2+ and Zn2+ (Figure 3 b).
4. Discussions
4.1 Family relationships of putative rice PCS/ PCS-like sequences
We investigated the relationship among these putative rice PCS/PCS-like sequences by generating an alignment of 14 identified PCS amino acid sequences followed by generation of a neighbor-joining phylogenetic tree (Figure 1a). The combined phylogeny PCS sequences revealed two subfamilies of putative orthologous genes,namely,OsPCS2,3,4,6,7,8,10,11,12,13 and 14; and OsPCS1,5,and 9,suggesting that PCS and PCS-like genes were divergent long years ago. OsPCS2,OsPCS 7 and OsPCS 14 have similar sequences to OsPCS11,OsPCS12,and OsPCS13,but are distributed in different 3 chromosomes. Other genes,OsPCS6 and OsPCS8,OsPCS3 and OsPCS4,are distributed in the same chromosome but with long evolutionary distance,and OsPCS1 and OsPCS9 are located in the same chromosome and are more closely related to each other (Figure 2 ),indicating that the divergence of genes was not relative to chromosome distribution.
4.2 The PCS genes might be divided into two subfamilies
Based on the phylogenetic tree of OsPCS genes,we inferred that phytochelatin synthase genes in eukaryotic organisms might be divided into two subfamilies,the PHYTOCHELATIN (PCS) subfamily serving as heavy metal chelators as formerly reported [31,32],and the unknown function gene family,namely,the PHYTOCHEL_SYNTH (PCS-like) subfamily.
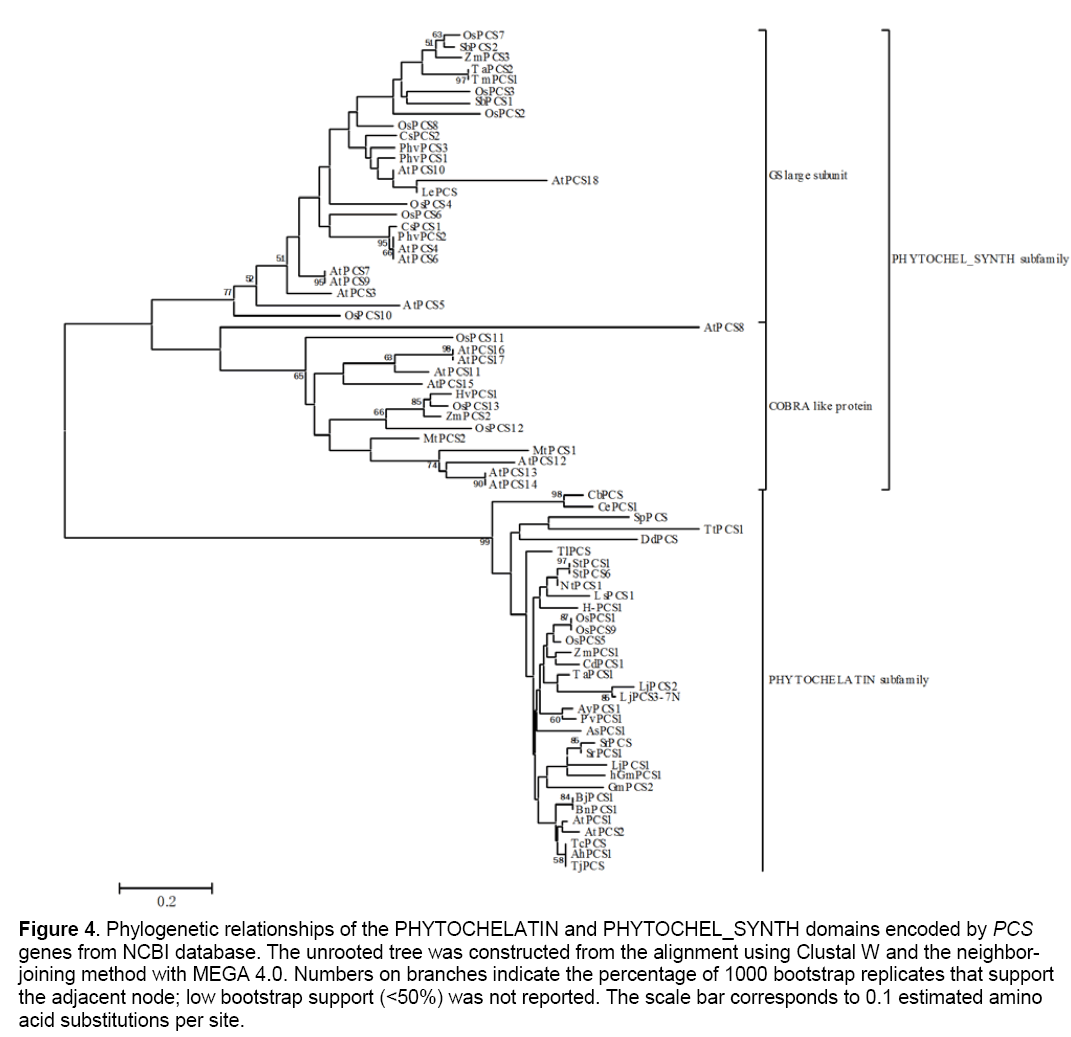
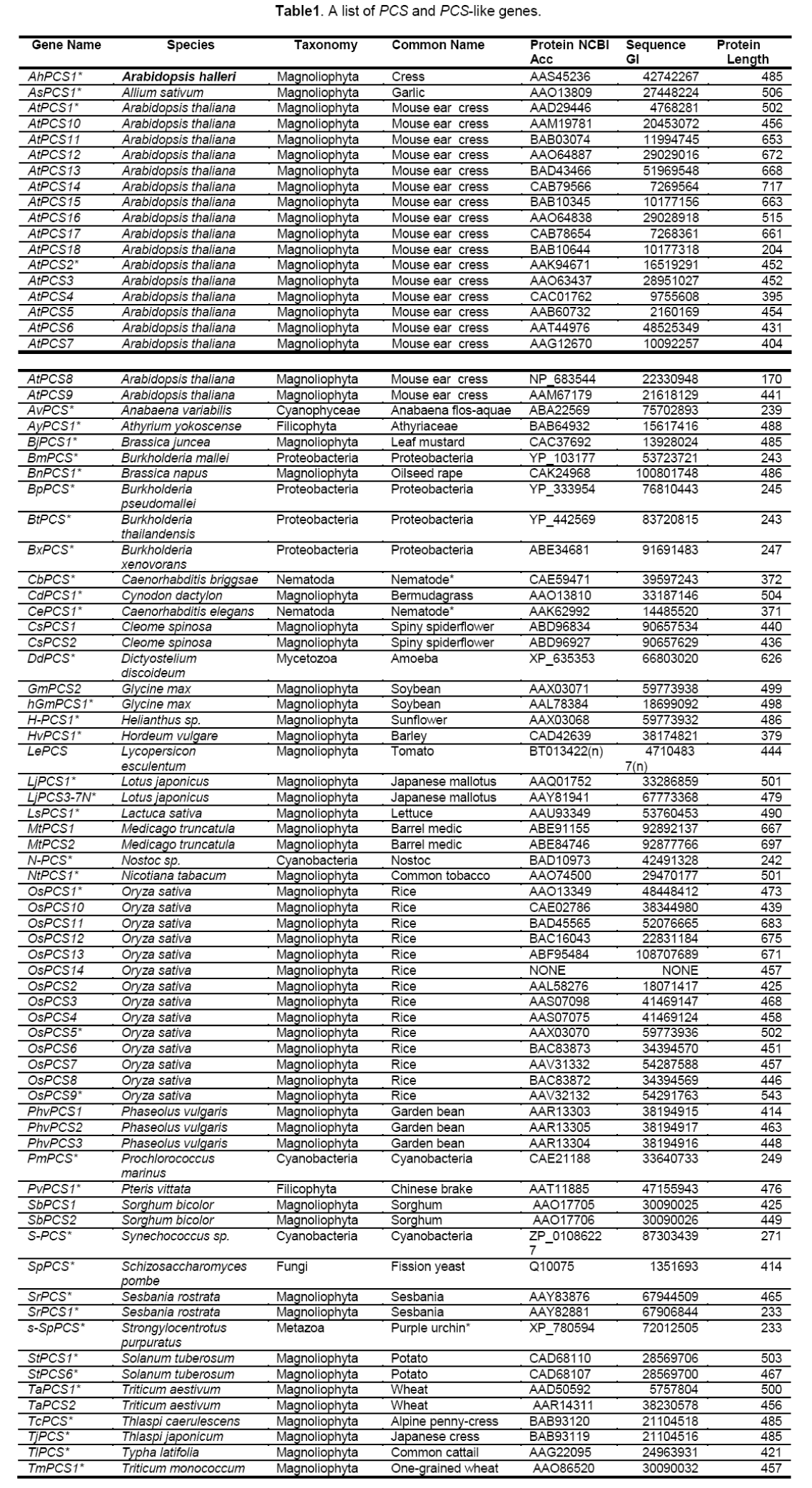
To validate our hypothesis,firstly,we searched in NCBI (www.ncbi.nlm.nih.gov) using the keywords “phytochelatin synthase,” and there were 94 items matched in protein database,22 in gene database and 115 in nucleotide database. Secondly,we searched in UniProtKB/Swiss-Prot (https://www.ebi. ac.uk/swiss-prot/) also using the keyword “phyto-chelatin synthase,” and there were 48 entries in protein database,and 157 in nucleotide database. Some retrieved sequences were discarded on the basis of the criteria described in Materials and Methods. All the remaining 87 sequences with information on species,sizes,and their accession numbers are available in Table 1. For the remote relationship between those family members,divergences take place not only in the peptide sequences but also in the nucleotide sequences,so it is very difficulty to put those subfamily members into a phylogenetic tree. We selected eukaryotic phytochelatin synthases to assess the evolutionary relationship between those subfamily members. Our results clear demonstrate that those family members can be classified into two subfamilies (Figure 4 ). To confirm our inference,we used BLAST to search all of the 87 sequences in NCBI,and found that 2 mono-domains,namely,the “PHYTO-CHELAYIN” and the “PYTOCHEL_SYNTH,” were matched. All of the “PHYTO- CHELAYIN” domain family members have conserved amino acids of Q,C,G,GH,P,and L in all organism species (see Supplemental Figure 2 a),while the “PYTOCHEL_ SYNTH” domain family is a C-rich family [28] with high probability in site of 49,69,95,and 149 in the full domain (see Supplemental Figure 2 b). Thus,phylogenetic relationships analysis of all predicted PCS genes from NCBI database clearly supports our hypothesis.
Figure 4: Phylogenetic relationships of the PHYTOCHELATIN and PHYTOCHEL_SYNTH domains encoded by PCS genes from NCBI database. The unrooted tree was constructed from the alignment using Clustal W and the neighbor-joining method with MEGA 4.0. Numbers on branches indicate the percentage of 1000 bootstrap replicates that support the adjacent node; low bootstrap support (<50%) was not reported. The scale bar corresponds to 0.1 estimated amino acid substitutions per site.
4.3 The PHYTOCHELATIN domain family diversity monogene to multi-gene family
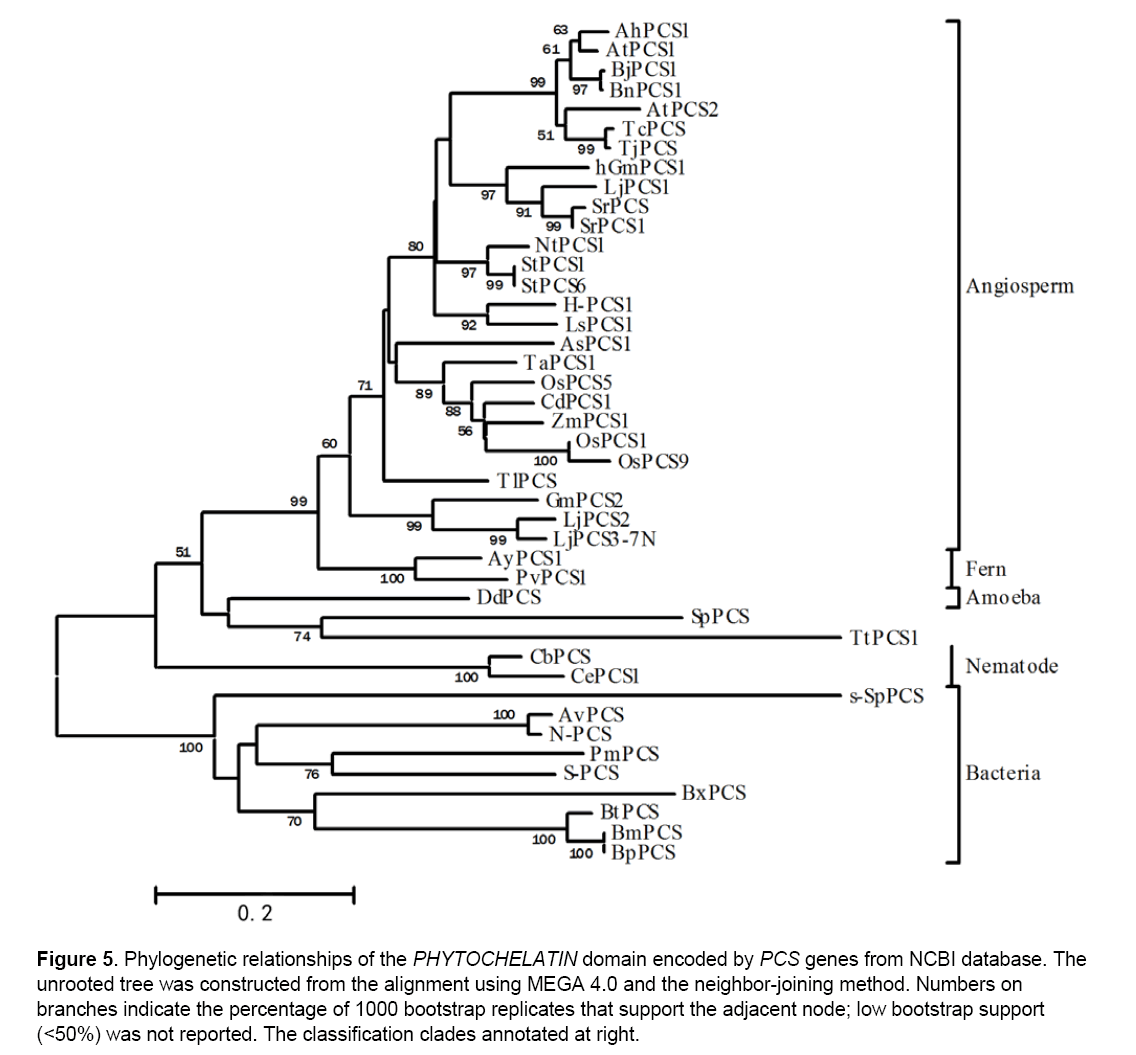
We used the mono-domain of Arabidopsis AtPCS1 to tBlastn and searched for “Phytochelatin” subfamily members in model organisms including all 30 species displayed in NCBI,and we also used tblastn to search in TIGR (www.tigr.org). We found that only 6 species (Dictyostelium discoideum,Schizosaccharomyces pombe,Canorhabditis elegans,Arabidopsis thaliana,Oryza sativa and Zea mays) contain the “PYTOCHEL_SYNTH” family genes. Interestingly,our result indicates that the genesis of “PYTOCHELATIN” gene subfamily might come from their mono-gene ancient ancestor,for in Dictyostelium discoideum,Schizosaccharomyces pombe and Canorhabditis elegans genomes only one gene was matched,while in Arabidopsis thaliana,Oryza sativa and Zea mays genomes more than one gene were matched (Figure 5 ). And the “PYTOCHEL_SYNTH” family might have evolved from the ancient ancestor of “PYTOCHELATIN” family.
Figure 5: Phylogenetic relationships of the PHYTOCHELATIN domain encoded by PCS genes from NCBI database. The unrooted tree was constructed from the alignment using MEGA 4.0 and the neighbor-joining method. Numbers on branches indicate the percentage of 1000 bootstrap replicates that support the adjacent node; low bootstrap support (<50%) was not reported. The classification clades annotated at right.
4.4 The bacteria PCS chelate heavy metal ions might not through Cys amino acids
Multi-alignment analysis of the functional domain of “PYTOCHELATIN” in bacteria showed that there was only one Cys in the conserved domain (Supplemental Figure 2 a),indicating that bacterial chelate heavy metal ions might have different mechanisms compared to eukaryotic ones.
5. Conclusions
Phytoremediation is an inovation area which interested more investigators who are concerned on heavy metal contamination. PCS involved critical roles in heavy metal remediation,predicating new PCS genes will inforce phytoremediation by transgenetic methods. And our findinds will interest those who are get in with phytoremediation.
Furthermore,the PCS-like family containing the cell wall protein COBRA-like and glutathione synthase (GS) large subunit (data not shown),will uncover the roles of COBRA-like and GS large subunit in heavy metal detoxification in the future.
Acknowledgments
This work was supported by grants from the Project of National Key Basic Research and Development of China (2007CB109305) and the National Natural Science Foundation of China (NO. 30671255).
References
- Cobbett C.S. (2000a) Phytochelatin biosynthesis and function in heavy-metal detoxification. Curr. Opin. Plant Biol.,3: 211-216.
- Cobbett C.S. (2000b) Phytochelatins and their roles in heavy metal detoxification. Plant Physiol.,123: 825-832.
- Li J.C.,Guo J.B.,Xu W.Z.,Ma M. (2007) RNA interference-mediated silencing of phytochelatin synthase gene reduce cadmium accumulation in rice seeds. J. Integr. Plant Biol.,49(7): 1032-1037.
- Vatamaniuk O.K.,Mari S.,Lu Y.P.,Rea P.A. (2000) Mechanism of heavy metal ion activation of phytochelatin (PC) synthase: blocked thiols are sufficient for PC synthase-catalyzed transpeptidation of glutathione and related thiol peptides. J. Biol. Chem.,275: 31451-31459.
- Piechalak A.,Tomaszewska B.,BaraÃÆââ¬Â¦Ãâââ¬Å¡kiewicz D. (2003) Enhancing phytoremediative ability of Pisum sativum by EDTA application. Phytochemistry,64(7): 1239-1251.
- Clemens S.,Kim E.J.,Neumann D.,Schroeder J.I. (1999) Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO J.,18(12): 3325-3333.
- Ha S.B.,Smith A.P.,Howden R.,Dietrich W.M.,Bugg S.,O'Connell M.J.,Goldsbrough P.B.,Cobbett C.S. (1999) Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe. Plant Cell,11: 1153-1164.
- Vatamaniuk O.K.,Mari S.,Lu Y.P.,Rea P.A. (1999) AtPCS1,a phytochelatin synthase from Arabidopsis: isolation and in vitro reconstitution. Proc. Natl. Acad. Sci. USA.,96: 7110-7115.
- Cazale A.C.,Clemens S. (2001) Arabidopsis thaliana expresses a second functional phytochelatin synthase. FEBS Lett.,507(2001): 215-219.
- Mizuno T.,Sonoda T.,Horie K.,Senoo K.,Tanaka A.,Mizuno N.,Obata,H. (2003) Cloning and characterization of phytochelatin synthase from a nickel hyperaccumulator Thlaspi japonicum and its expression in yeast. Soil Sci. Plant Nutr.,49: 285-290.
- Oven M.,Page J.E.,Zenk M.H.,Kutchan T.M. (2002) Molecular characterization of the homo-phytochelatin synthase of soybean Glycine max: relation to phytochelatin synthase. J. Biol. Chem.,277: 4747-4754.
- Tsuji N. ,Nishikori S.,Iwabe O.,Shiraki K.,Miyasaka H.,Takagi M.,Hirata K.,Miyamoto K. (2004) Characterization of phytochelatin synthase-like protein encoded by alr0975 from a prokaryote,Nostoc sp. PCC 7120. Biochem. Biophys. Res. Comm.,315(3): 751-755.
- Dong R. (2005) Molecular cloning and characterization of a phytochelatin synthase gene,PvPCS1,from Pteris vittata L. J. Ind. Microbiol. Biot.,32(11-12): 527-533.
- He Z.Y.,Lia J.C.,Zhang H.Y.,Ma M. (2005) Different effects of calcium and lanthanum on the expression of phytochelatin synthase gene and cadmium absorption in Lactuca sativa. Plant Sci.,168(2): 309-318.
- Loscos J.,Naya L.,Ramos J.,Clemente M.R.,Matamoros M.A.,Becana M. (2006) A reassessment of substrate specificity and activation of phytochelatin synthases from model plants by physiologically relevant metals. Plant Physiol.,140: 1213-1221.
- Sauge-Merle S.,Cuine S.,Carrier P.,Lecomte-Pradines C.,Luu D.T.,Peltier G. (2003) Enhanced toxic metal accumulation in engineered bacterial cells expressing Arabidopsis thaliana phytochelatin synthase. Appl. Environ. Microbiol.,69(1): 490-494.
- Sriprang R.,Hayashi M.,Ono H.,Takagi M.,Hirata K.,Murooka Y. (2003) Enhanced accumulation of Cd2+ by a Mesorhizobium sp. transformed with a gene from Arabidopsis thaliana coding for phytochelatin synthase. Appl. Environ. Microbiol.,69(3): 1791-1796.
- Konishi T.,Matsumoto S.,Tsuruwaka Y.,Shiraki K.,Hirata K.,Tamaru Y.,Takagi M. (2006) Enhancing the tolerance of zebrafish (Danio rerio) to heavy metal toxicity by the expression of plant phytochelatin synthase. J.Biotech.,122(3): 316-325.
- Pomponi M.,Censi V.,Di Girolamo V.,De Paolis A.,Di Toppi L.S.,Aromolo R.,Costantino P.,Cardarelli M. (2006) Overexpression of Arabidopsis phytochelatin synthase in tobacco plants enhances Cd2+ tolerance and accumulation but not translocation to the shoot. Planta,223(2): 180-190.
- Lee S.,Moon J.S.,Ko T.S.,Petros D.,Goldsbrough P.B.,Korban S.S. (2003) Overexpression of Arabidopsis phytochelatin synthase paradoxically leads to hypersensitivity to cadmium stress. Plant Physiol.,131: 656-663.
- Li Y.,Dhankher O.P.,Carreira L.,Lee D.,Chen A.,Schroeder J.I.,Balish R.S.,Meagher R.B. (2004) Overexpression of phytochelatin synthase in Arabidopsis leads to enhanced arsenic tolerance and cadmium hypersensitivity. Plant Cell Physiol.,45(12): 1787-1797.
- Jiang S.Y.,Jasmin P.X.,Ting Y.Y.,Ramachandran S. (2005) Genome-wide identification and molecular characterization of Ole_e_I,Allerg_1 and Allerg_2 domain-containing Pollen-Allergen-like genes in Oryza sativa. DNA Res.,12(3): 167-179.
- Yoshida S.,Forno D.A.,Cock J.H. (1971) Laboratory manual for physiological studies of rice. Manila,The Philippines: International Rice Research Institute.
- Reale S.,Doveri S.D.,Angiolillo,A.,Lucentini L.,Pilla F.,Martín A.,Donini P.,Lee D. (2006) "SNP-based markers for discriminating olive (Olea europaea L.) cultivars. (single- ucleotide polymorph",Genome,49(9): 1193-1205.
- Cai X.J.,Lytton J. (2004) The cation/Ca2+ exchanger superfamily: phylogenetic analysis and structural implications. Mol. Biol. Evol.,21(9): 1692-1703.
- Tamura K.,Dudley J.,Nei M.,Kumar S. (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol.,24(8): 1596-1599.
- Saitou N.,Nei M. (1987) The neighbor-joining method: a new method for reconstruction of phylogenetic trees. Mol. Biol. Evol.,4: 406-425.
- Schuster-Böckler B.,Schultz J.,Rahmann S. (2004) HMM Logos for visualization of protein families. BMC Bioinformatics,5: 7.
- Linding R.,Russell R.B.,Neduva V.,Gibson T.J. (2003) GlobPlot: exploring protein sequences for globularity and disorder. Nucl. Acids Res.,31(13): 3701-3708.
- Altschul S.F.,Madden T.L.,Schäffer A.A.,Zhang J.,Zhang Z.,Miller,W.,Lipman D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res.,25(17): 3389-3402.
- Cobbett C.S.,Goldsbrough P. (2002) Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Ann. Rev. Plant Biol.,53: 159-182.
- Rea P.A.,Vatamaniuk O.K.,Rigden D.J. (2004) Weeds,worms,and more. Papain's long-lost cousin,phytochelatin synthase. Plant Physiol.,136: 2463-2474.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences