Discovery of ÃÆà ½Ãâò -lactam Antibiotic Resistance Specific Functional Residues: A Bioinformatics Approach
Rajendra Mandage, Padmaja Kamath, Monali Wakle, Afaque Momin
Rajendra Mandage*,Padmaja Kamath,Monali Wakle,Afaque Momin
Dept. of Bioinformatics,GNIRD,G. N. Khalsa College,Matunga,Mumbai,400019 India.
Abstract
Since the introduction of β-lactam antibiotics, disease causing microbial resistance to these antibiotics has become a problem increasingly. Discovery of functional residues in beta-lactamase that play major role in antibiotic resistance provide an opportunity to understand their fundamental molecular mechanisms. We present an example of extraction of functional information from protein 3D structures using a bioinformatics approach in the context of the antibiotic resistance. In case studies, 45 homologous beta-lactamase sequences were investigated using a homology based approach by ConSurf server to analyse the surface of beta-lactamase to reveal common functional features which might facilitate them to identify lactam antibiotics. We have identified functional residues using phylogenetic studies, protein sequence MSA and three-dimensional mapping. The results demonstrate the presence of antibiotic resistance specific highly conserved residues comprising of high proportion of surface-exposed hydrophobic residues as do not endure amino acid substitutions, signifying that they have critical role in antibiotic resistance and the remaining positions tolerate amino acid substitutions and may affect the substrate specificity of the beta lactamase. These functionally important residues could also potentially be used in the rational design of novel, efficient antimicrobial agents
Keywords
beta-lactamase; drug resistance; 3D structure; functional residues; ConSurf server; PSI-BLAST.
1. Introduction
Lactam antibiotics,including penicillins and cephalosporins,are among the most prevalent antibacterial agents administered worldwide. The most common bacterial resistance mechanism against these antibiotics is the production of lactamase enzymes [1],which hydrolyze the beta-lactam bond,rendering the antibiotics inactive. As beta-lactam resistance has emerged as a significant public health risk,so it would be interesting to understand the characteristics of beta-lactamases at molecular level to aid the design of new classes of antibiotics.
The three-dimensional structure of a complete beta-lactamase has been recently determined (pdb id 3blm). In this work,we have used this structural information for identification of functional residues in beta-lactamase by structure-mapping of phylogenetic information through a computer-based structure approach. Protein function is an outcome of the three dimensional organization adopted by the protein sequence and therefore a protein’s 3D structure may be more suitable to identify functional residues because they probe directly the molecular basis of function through small structural motifs of just a few key amino acid that,ideally,identify functional determinants and their functionally relevant matches in other proteins [2-9]. Hence,referring to functional residues for protein function includes both types of residues: residues functional for protein structure and/or residues functional for its biological function (e.g.,catalysis,binding). When a sufficient amount of homolog are detected,powerful prediction schemes can be based on the observation that evolutionarily conserved regions are often functionally important,typically,only the principal functionally important region of the protein is detected,is a functional step in the study of the biological function of proteins [10]. This task is especially important for proteins with a known structure and unknown function. Detection of key amino acid positions that are functionally important is also essential for drug design studies,for protein classification and annotation and for evolutionary studies [11]. In an attempt to identify the functional residues of beta lactam antibiotics,we used the ConSurf server (https://consurf.tau.ac.il/) which enables identification of functionally important regions on the surface of a protein or domain,based on the phylogenetic relations between its close sequence homologues,and projects the data onto a representative crystal structure [12].
2. Methods
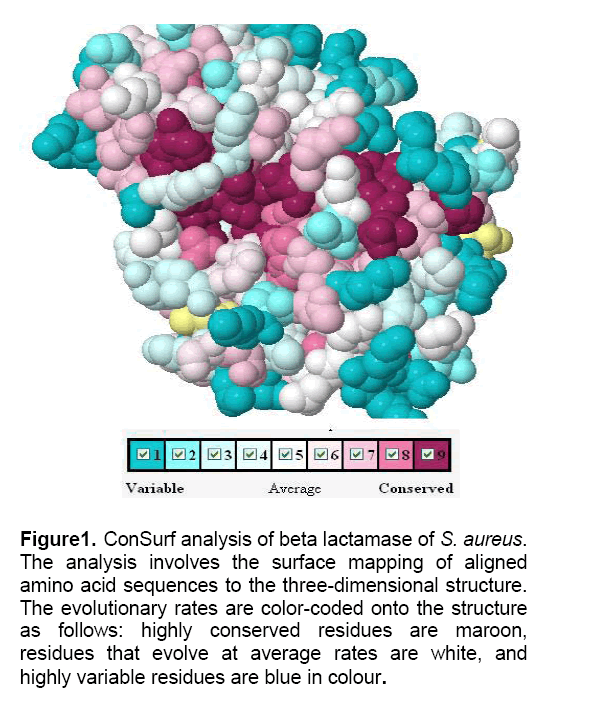
The reference beta-lactamase protein structure (pdb id 3blm) was retrieved from the pdb based on keyword search and used to screen the SWISS-PROT database for homologous proteins through PSI-BLAST [13] analysis with E-value cut off of 0.001 using the ConSurf web server). This is an algorithmic tool for the identification of functionally important regions in proteins by estimating the degree of conservation of the amino-acid sites among their close sequence homologues. It first aligns amino acids sequences by ClustalW algorithm with default parameters; then builds phylogenetic tree using the Neighbor-Joining method; and finally calculates conservation scores for each specific position amino acid residues by means of an empirical Bayesian algorithm; Amino acid residues were clustered into average,variable and conserved scale for three-dimensional visualization purpose. Lactamase protein is represented as a space fill model,where the residue conservation scores are color-coded onto its Van der Waals surface. The color-coding bar shows the coloring scheme; conserved amino acids are colored bordeaux,residues of average conservation are white,and variable amino acids are turquoise.
3. Results and Discussion
The Cornsurf server detected 45 homologous proteins by PSI-blast analysis against Swissprot database that showed low e-values (3e-42 to e-100) and high identity (40-95%) with other beta-lactamase proteins. These proteins were from various bacteria obtained through evolutionary scale,including Staphylococcus aureus,Bacillus cereus,Brevibacillus brevis,Micromonospora sps,Streptomyces flavogriseus,Clostridium botulinum,Pseudomonas luteola and many more. The selected protein sequences were examined for common residue patterns using the ConSurf server,searching for common antibiotic resistance-specific residues. To gain insights into the relative position of conserved residues,we analyzed the three-dimensional structure of beta lactamase using a nine grade color scale to represent the conservation scores of each amino acid as implemented in the ConSurf server. Antibiotic resistance-specific and accessible residues that are either highly conserved (levels 8–9,bordeaux colour,) or highly variable (levels 1–2,blue colour),but which have only average scores (white) in homologous beta lactamase sequences,are underlined in Figure 1.
Figure 1: ConSurf analysis of beta lactamase of S. aureus. The analysis involves the surface mapping of aligned amino acid sequences to the three-dimensional structure. The evolutionary rates are color-coded onto the structure as follows: highly conserved residues are maroon, residues that evolve at average rates are white, and highly variable residues are blue in colour.
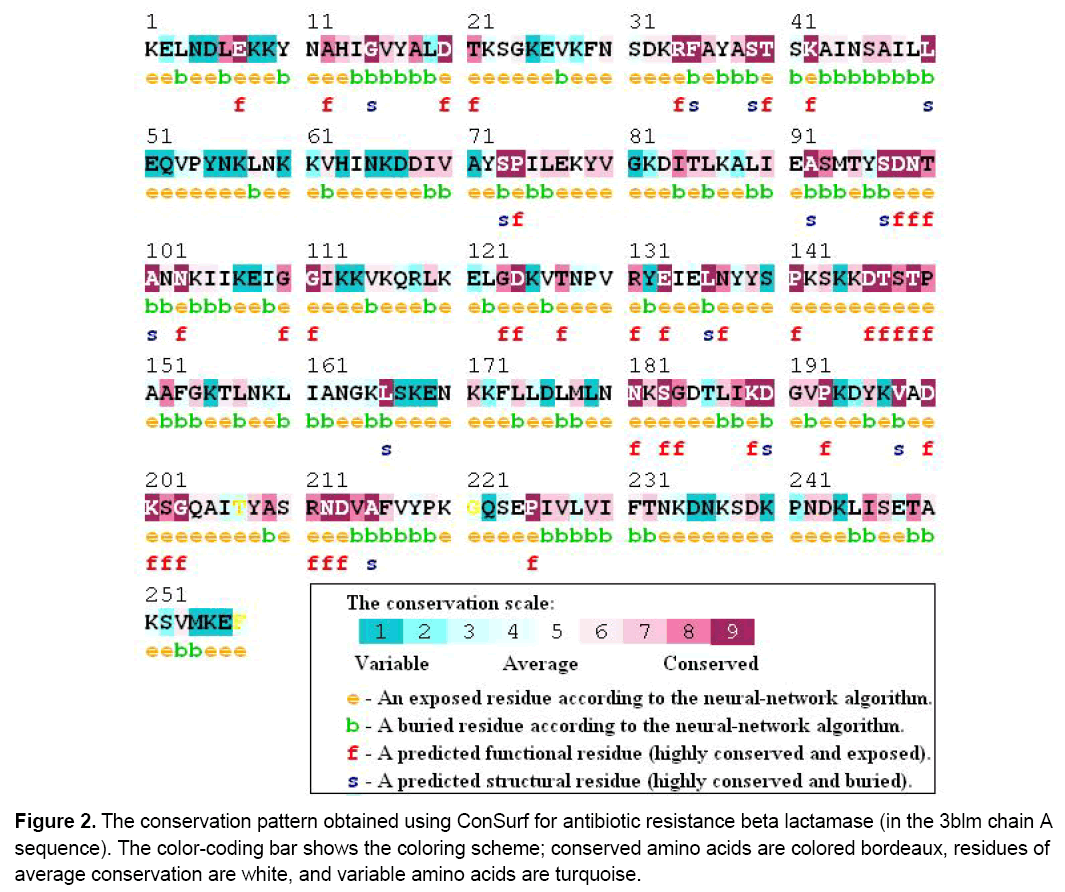
Results showed that ten amino acids were in the ninth grade,indicating that they are the most conserved of the predicted conserved residues Figure 2 . 38 of them were highly conserved in all homologous proteins (E7, G15, D20, R34, F35, S39, T40, K42, L50, S73, P74, A92, S97, D98, N99, A101, N103, G111, D124, E133, L136, P141, D146, T147, T149, L166, N181, S183, K189, D190, P193, V198, D200, K201, G203, N212, D213, A215, P225).
Figure 2: The conservation pattern obtained using ConSurf for antibiotic resistance beta lactamase (in the 3blm chain A sequence). The color-coding bar shows the coloring scheme; conserved amino acids are colored bordeaux, residues of average conservation are white, and variable amino acids are turquoise.
Figure 2 do not tolerate residue substitutions and therefore make critical contributions to enzyme structure and function whereas the others were substituted residues including (K/G)95,(G/K)103,(I/M)106,(G/A)109,(K/P)111,(A/S/T)112 and (G/Y)157. Most of the residues known to be involved in the catalytic mechanism and in substrate binding are conserved,In particular,we identified position the consensus sequences 146DTSTA150,201KSG203,and 110GGP112 as the most likely candidates for determination of antibiotic specificity. In total,38 residues were observed to be essential for functionality. This indicates that the active site of beta lactamase is much optimised for antibiotic resistance therefore it is in general intolerant of residues substitutions. We suggest that our new approach for functional residues identification not merely complements to other well-known methodologies but also provides a deeper understanding of an evolution a 3D structure of proteins and their specific functional residues. This relationship is probably a direct consequence of the necessary functional robustness of proteins.
Transforming protein structural information into functional information permitted us to inspect the connection between each of functional amino acid residues and the molecular mechanism as a whole. Such functional residues might interact with most other residues directly or by a few intermediates to assist molecular mechanism. The ConSurf server helps in the discovery of such key functional residues on the surface of beta lactamase 3D structure based on the phylogenetic relations between its close sequence homolog. It was able to successfully discover homolog sequences from protein sequence database. There could be different possible strategies exist which may provide essential mechanism to accomplish exquisite antibiotic resistance. First is that the functional residues may be shaped by set of contiguous conserved and variable residues,which,are mostly hydrophobic nature results in position-specific effects that direct catalysis of antibiotic. Second,functional residues of beta lactamase may have evolved to resist beta lactum antibiotics consisting of a chain of hydrophobic amino acids,rather than depending on the exact sequence amino acid. Since the 45 sequences studied here constitute only a small sample of beta lactamase,it may be premature to generalize this outcome. However,this is the first demonstration of antibiotic resistance-specific surface feature displayed by structural and functional aspects of sequences of the beta lactamase. Identification of such functional residues will give an accurate and deep intuitive understanding for target selection and efficiently focusing mutational studies on their key residues,thus concerning raw sequence and structure data to functional information. It could also potentially be used in the rational design of novel,efficient antimicrobial agents.
4. Conclusions
The recent deposition of experimentally determined 3D structure of protein along with insilico model structure from amino acid sequence has resulted in an increased significance of computer-based methods for protein function prediction. Now our potential to decode these large amounts of structural information into functional information at the molecular level must depend on bioinformatics approaches. Clearly,evolving lactamase causes a serious threat to antimicrobial treatment and poses challenge to existing use of antibiotics. Therefore,the detailed depiction of such protein is essential for the design of novel lactamase inhibitors to combat resistance problem. Our results demonstrated that the use of CornSurf server can predict the relative intolerance of lactamase for amino acid substitutions may have important implications for the evolution of antibiotic resistance. This is helpful to design targeted mutagenesis analysis and that can used as a platform to interpret outcomes in the broad context of antibiotic resistance. Bioinformatics based discovery of such functional residues greatly boost our ability to make a great progress in drug design,protein engineering through site-directed mutagenesis,and in depth functional annotation.
References
- Wallace AC.,Borkakoti N.,Thornton JM. (1997) TESS: a geometric hashing algorithm for deriving 3D coordinate templates for searching structural databases. Application to enzyme active sites. Protein Sci,6:2308–2323.
- Barker JA.,Thornton JM. (2003) An algorithm for constraint-based structural template matching:application to 3D templates with statistical analysis. Bioinformatics,19:1644–1649.
- Kleywegt GJ. (1999) Recognition of spatial motifs in protein structures. J Mol Biol,285:1887–1897.
- Stark A.,Russell RB. (2003) Annotation in three dimensions. PINTS: Patterns in Non-homologous Tertiary Structures. Nucleic Acids Res,31:3341–3344.
- Artymiuk PJ.,Poirrette A.R.,Grindley H.M. et al. (1994) A graph-theoretic approach to the identification of three-dimensional patterns of amino acid side-chains in protein structures. J Mol Biol,243:327–344.
- de Rinaldis M.,Ausiello G.,Cesareni G. et al. (1998) Three-dimensional profiles: a new tool to identify protein surface similarities. J Mol Biol,284:1211–1221.
- Laskowski RA. (1995) SURFNET: a program forvisualizing molecular surfaces,cavities,and intermolecular interactions. J Mol Graph,13:323–330. 307–8.
- Kleywegt GJ.,Jones TA. (1994) Detection,delineation,measurement and display of cavities in macromolecular structures. Acta Crystallogr D Biol Crystallogr,50 :178–185.
- Lichtarge O.,Sowa ME. (2002 Feb) Evolutionary predictions of binding surfaces and interactions.Curr Opin Struct. Bio,12: 21-27.
- Fabian Glaser.,Tal Pupko.,Inbal Paz. (2003) ConSurf: Identification of Functional Regions in Proteins by Surface-Mapping of Phylogenetic Information. Bioinformatics,19: 163–164.
- Rost,B.(2002) Enzyme function less conserved than anticipated. J. Mol. Biol.,318: 595–608.
- Ruta Furmonaviciene,Brian J. Sutton,Fabian Glaser,(2005) An attempt to define allergen-specific molecular surface features: a bioinformatic approach. bioinformatics .,21: 4201–4204
- Altschul,S.F. et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res,25: 3389–3402.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences