Considering the Effects of Ultra Violet Treated Acidithiobacillus sp. on the Expression of rus Operon Genes at Uranium Bioleaching
Samaneh Jahani, Faezeh Fatemi*, Saba Miri
Samaneh Jahani1, Faezeh Fatemi2*, Saba Miri3
1Department of Microbiology, Faculty of Science, Qom Branch, Islamic Azad University, Qom, Iran
2Materials and Nuclear Fuel Research School, Nuclear Science and Technology Research Institute, Tehran, Iran
3Department of Biotechnology, Faculty of Life Science, Alzahra University, Tehran, Iran
Received date: November 28, 2018; Accepted date: January 15, 2019; Published date: January 25, 2019
Citation: Jahani S, Fatemi F, Miri S. Considering the Effects of Ultra Violet Treated Acidithiobacillus sp. on the Expression of rus Operon Genes at Uranium Bioleaching. Electronic J Biol, 15:1
Abstract
Acidithiobacillus sp. obtains energy from the oxidation of Fe2+ or reduced sulfur compounds. The proteins encoded by the rus operon are involved in the electron transfer from Fe2+ to O2. In this research, the expression of rus operon, proposed to evaluate the effects of pulp densities and UV-mutations in the Acidithiobacillus sp. FJ2 in the bioleaching process. As observed by real-time PCR, the rus operon genes were affected by pulp density and UV-mutations. rus operon gene expressions were modified differently by pulp density while, a number of induced bacteria were expressed more than control. Furthermore, at 5%, 10% and 50% pulp densities, the extraction of uranium by UV-induced bacteria was higher than that by the control bacteria. These results indicated that UV-mutations could affect the bacterial activity at bioleaching process which strongly supports the idea to facilitate the involvement of the rus operonencoded products in the oxidation of ferrous iron, leading to ore uranium extraction.
Keywords
Acidithiobacillus sp.; FJ2; rus operon; UV-mutation; Uranium bioleaching
1. Introduction
Recently, some bacteria, which can grow in the extreme condition, have attracted significant attention due to their physiology and ecology. Among them, Acidithiobacillus ferrooxidans is considered to play the most main role in leaching of sulfide minerals [1]. This bacterium is a Gram-negative eubacterium, acidophil and chemolithotroph. It is able to do the biological oxidation of ferrous iron to ferric iron which it is a powerful oxidant agent to oxidizing insoluble metal sulfides to their soluble form. This process is known as bioleaching that used for recovery of several metals such as copper, uranium and gold from low grade ores [2].
The respiratory chain of A. ferrooxidans follows the pathway: Fe(II) —»Cyc2 —»rusticyanin —»Cyc1 —»cytochrome oxidase aa3 (Cox BACD) —»O2 cytoplasmic [3]. rus operon encoded these proteins, which are organized in the A. ferrooxidans genome in the following: cyc2, cyc1, ORF1, coxB, coxA, coxC, coxD, and rus. A c-type cytochrome encode by cyc2 gene (outer membrane) [3], a cytochrome c4 encode by cyc1 gene (periplasmic), and a putative outer membrane protein encode by ORF1 [4]. The subunits of an aa3-type cytochrome oxidase encode by coxBACD genes (inner membrane), and a periplasmatic blue copper protein (rusticyanin) encode by rus gene [5].
Bioleaching process has relatively incompetent because of their low cell densities, slow growth rate, inhibition of iron oxidation by ferric iron and their low activity in presence of high pulp density of sulfide ore [6]. Such problems demand the improvement of this process.
Recently, we optimized the bioleaching process at high pulp density regarding the ecological parameters (pH, redox potential, temperature, rpm, inoculum, concentration of FeSO4.7H2O and (NH4)2SO4) using isolated Acidithiobacillus sp. FJ2 [7,8]. In followings, we decided to consider the bacteria modulation effects in the bioleaching improvement process via UV-mutations. The UV-induced mutagenesis is a regularly used and effective method for improvement of the bacteria. Many studies on the mutation of bioleaching bacteria have been done all over the world, but no report has focused on the evaluation of relative rus operon gene expressions of UVinduced bacteria at the presence of uranium ore pulp density variations. So, in this paper, we analyzed quantitatively the expression of four genes from the rus operon when UV-induced Acidithiobacillus sp. FJ2 was grown in the presence of variable uranium ore pulp densities in the achievement of 100% uranium extraction rate.
2. Materials and Methods
2.1 Microorganism and growth conditions
Acidithiobacillus sp. FJ2 isolated from sulfur spring of Ramsar, Iran, was cultured in 9k medium [8]. The pH of 9k medium was adjusted to 2.0 with 10N H2SO4. The flask was then placed on constant-temperature shakers at a rotation speed of 180 rpm at 30°C until the color of the medium became reddish-brown.
2.2 UV-mutation of strain
The FJ2 strain in the logarithmic growth phase was centrifuged for 20 min at 4000 rpm, and the cells were obtained with removing the supernatant. After that, the cells were washed three times using acidified distilled water with pH=2. Finally, the cells were suspended in salts medium without energy source and the density of the cells was adjusted to 1×108 cell mL-1 [9]. In following, 10 mL of pre-cultured suspension of the strain was transferred to 4 plates. A distance of 30 cm was kept between the plates and UV light. The isolate were UV irradiated for periods of 0, 60, 120 and 180 s, respectively at the power of 30 W and wavelength of 254 nm. After mutation, the samples were kept away from the light and stored in the refrigerator at 4°C for 12 h to prevent the bacteria from recovery with light [10]. After mutation, the bacterial mutants were cultivated in 9K liquid medium under optimal growth conditions for using in the uranium bioleaching experiments.
2.3 Bioleaching experiments
Bioleaching experiments were carried out in 2000 mL flasks containing 900 mL optimized 9k medium containing: (NH4)2SO4 2.0 g/L, K2HPO4 0.5 g/L, MgSO4.7H2O 0.5 g/L, KCl 0.1 g/L, Ca(NO3)2 0.01 g/L, FeSO4.7H2O 20 g/L, peptone 0.5 g/L, tryptic soy broth (TSB)1.0 g/L, yeast extract 0.01% [11] (pH=2) and 100 mL of control and induced bacteria. A low-grade uranium ore sample was prepared from Saghand mine (Anomaly NO.2) of Yazd in south of Iran (d80=106 μm). The chemical composition analysis results of ore sample are shown in Table 1. X-ray Diffraction (XRD) results showed that uranium ore sample is mainly composed of Pyrite (FeS2) and Uraninite (UO2). Various amount of uranium ore powder was added into the flasks containing media to the final concentration of 5%, 10%, 15%, 25% and 50% (w/v), respectively. In addition, the negative control experiment was carried out by addition of 100 mL methanol-formaldehyde solution (10:1) to the medium instead of inocula. Finally, the flasks were maintained at 35°C and shaken at 150 rpm. During bioleaching, pH value, redox potential, ferrous iron and uranium ion concentrations in the leaching solution were determined at certain intervals. The loss of water due to evaporation was compensated by acidified distilled water with pH=2 in order that the solution was maintained at 1000 mL in the flasks [12].
| W(S)/% | W(Fe2O3)/% | ppm(U) | W(P2O5)/% |
|---|---|---|---|
| 0.75 | 49.67 | 465 | 0.23 |
| W(Al2O3)/% | W(MgO)/% | W(V2O5)/% | W(SiO2)/% |
| 1.49 | 19.03 | 0.37 | 24.83 |
Table 1: Chemical compositions of the uranium ore.
2.4 Analytical methods
Ferrous ion was determined by the colorimetric measurement of formed red-colored ferric– sulfosalicylate complex that described by Golmohammadi et al. [13]. The concentration of dissolved uranium ion in the leaching solution was analyzed by Inductively Coupled Plasma-Mass Spectrometry (ICP-MS) technique. The pH value and redox potential (Eh) were measured by a pH/Eh process controller (Metrohm, model 827).
2.5 Preparation of samples for real-time PCR
At the end of the bioleaching experiments (extracted uranium was 100%), the cells of control and induced samples in the leach-liquor were centrifuged for 20 min (4000 rpm) and suspended in 110 mL 9k salts medium without Fe2+. 100 mL of the resulting suspensions of bacteria from lower pulp density were inoculated into bioleaching soluble with higher pulp density as inocula [14]. The remaining suspensions of bacteria were used for RNA extraction process.
2.6 RNA extraction and cDNA generation
Total cellular RNA was isolated using the GeneJET RNA purification kit (Thermo Scientific) according to the manufacturer’s protocol. RNA extracts were treated with Thermo Scientific DNase I (RNase-free), to remove genomic DNA. Then, total cellular RNA was quantified at OD260 and OD280 with a NanoDrop 2000 spectrophotometer (Thermo Scientific). The purified RNA from each sample served as the template to generate cDNA with RevertAid first strand cDNA synthesis kit (Thermo Scientific) and Random Primers (Thermo Scientific). Eventually, PCR was carried out for process correctness.
2.7 Quantitative real-time PCR analysis
In this study, the relative rus operon gene expressions (cyc2, rus, cyc1 and coxB) of control and induced Acidithiobacillus sp. FJ2 in the presence of 5%, 10%, 15%, 25% and 50% of uranium ore were determined using real-time PCR method. Primers used in this study were present in Table 2.
| Name | Sequence (5'–3') |
|---|---|
| Cyc2(F) Cyc2(R) |
CCGCCAGAGTAGGTCAAATGC AACTCTAATGCGGGTGCTTCTC |
| Rus(F) Rus(R) |
GGCATAACCGCATAAGGAGGT GAACCCGACCTTGGAGATTCC |
| Cyc1(F) Cyc1(R) |
TTCTGGGCGTTGAAGTAATCCG TGAAAGCGTATAAGGACCACTCC |
| CoxB(F) CoxB(R) |
GCTCCCTATCTGGTCAAACAACT CGTGATCCCAAATGAAGTGCG |
| 16S rRNA(F) 16S rRNA(R) |
CCTACGGGAGGCAGCAG CGGTGCTTCTTCTTGGATTCACG |
Table 2: Nucleotide sequence of the primers for real-time PCR.
All real-time PCR mixture (final volume 10 μL) contained 5 μL of SYBR Green Real-time PCR Master Mix (Takara Premix Ex Taqkit), 0.2 μL of a 10 mM primers, 5 μL of cDNA template, 0.2 μL of Rox, and 3.9 μL of nuclease-free water.
The real-time PCR was carried out with a step one Real-Time PCR System (Applied Biosystems): 1 cycle of 95°C for 30 s, and then 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 20 s. At the completion of each run, melting curves for the amplicons were measured by raising the temperature by 0.3°C from 55 to 95°C while monitoring fluorescence. The specificity of the PCR amplification was checked by examining the derivative melting curve for Tm, its symmetry and the lack of non-specific peaks. Three parallel measurements for each cDNA sample from independent RNA isolation were detected. The genes expression ratio was recorded as the fold difference in quantity from control and mutated samples that were in the presence of various uranium pulp densities. The results were normalized against house-keeping gene 16S rRNA to correct the sample-to-sample variation [15].
2.8 Statistical analysis
Data are presented as means ± Standard Error of Mean (SEM) of three samples (triplicate). The results were subjected to one-way ANOVA followed by Tukey’s HSD using SPSS (version 22) software. Significant levels were defined as p<0.05.
3. Results
3.1 Uranium extraction yields at the control and induced bacteria in the uranium bioleaching processes
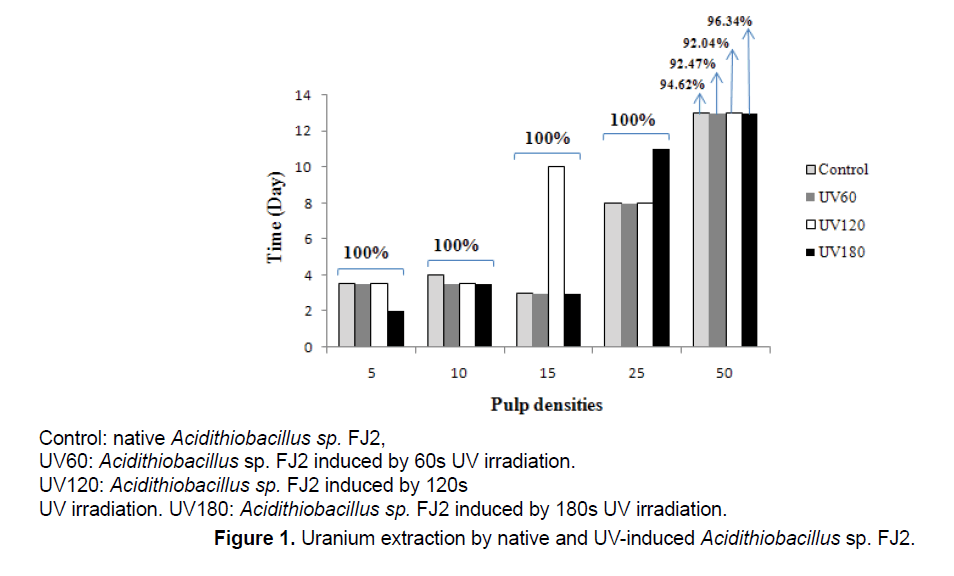
Figure 1 shows the effects of different pulp densities on the amount of uranium leached from the low-grade ore by control and induced Acidithiobacillus sp. FJ2. The results indicated that, with increasing amounts of ore in the medium, the yields of biologically extracted uranium were decreased.
During the bioleaching process at 5%, 10% and 50% pulp densities, the extraction of uranium by UVinduced bacteria was higher than that by the control bacteria. In leaching process at 5% pulp density, by the UV180 sample, the maximum leaching of uranium reached to 100% after 2 days, while for the another samples was 3 days. At 10% pulp density, all mutants’ bacteria were more effective than the control sample. In addition, at 50% pulp density, the highest uranium extracted was observed in all samples, which indicates that 96.34% uranium solubilization in UV180 during 13 days. When Acidithiobacillus sp. FJ2 induced by 60 and 120 s of UV, it was observed that the uranium solubilizations were by 92.47% and 92.04%, respectively. On the other hand, the control sample (the bacterium without mutant), was able to do 94.62% uranium extraction during 13 days at the same pulp density. According to the results, the best mutation effect of the strain for the solubilization of uranium is UV180 s.
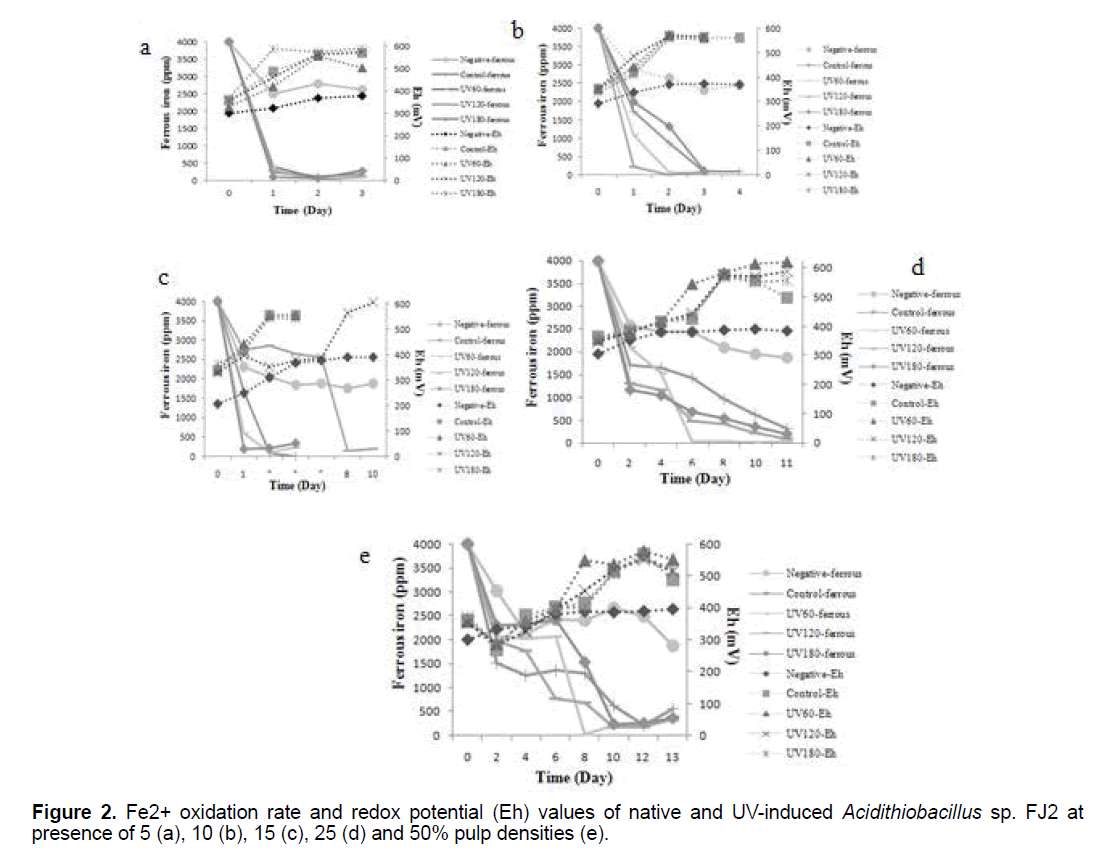
3.2 Ferrous bio-oxidations and redox potentials in the uranium bioleaching processes
The effects of variable pulp densities on ferrous bio-oxidations and redox potentials of control and induced bacteria are shown in Figure 2. As can be seen, the soluble ferrous was decreased, while the redox potential gradually increased in all the samples (p<0.05). These profiles indicate the bacterial activity as a result of increased the leached process. From these results, it could be deduced that at lower pulp density, the rate of ferrous oxidation to ferric was faster than the higher pulp density. Hence, with increasing of pulp density, more time is needed for iron oxidation process that this pattern was observed in all samples clearly.
In the case of UV180 at 5% pulp density, the redox potential (Eh) began to increase rapidly from 369 mV to about 590 mV significantly after 3 days (p<0.05). While, the redox potentials increased up to 563-567 mV in control sample at the same time. In addition, at 15% pulp density, increase in redox potential and reduce in amount of ferrous ion were observed in presence of UV120 sample (Figure 2c). Besides, it is clear from the Figure 2, the lowest values of redox potential are observed in the negative tests curves, without bacterial inoculum.
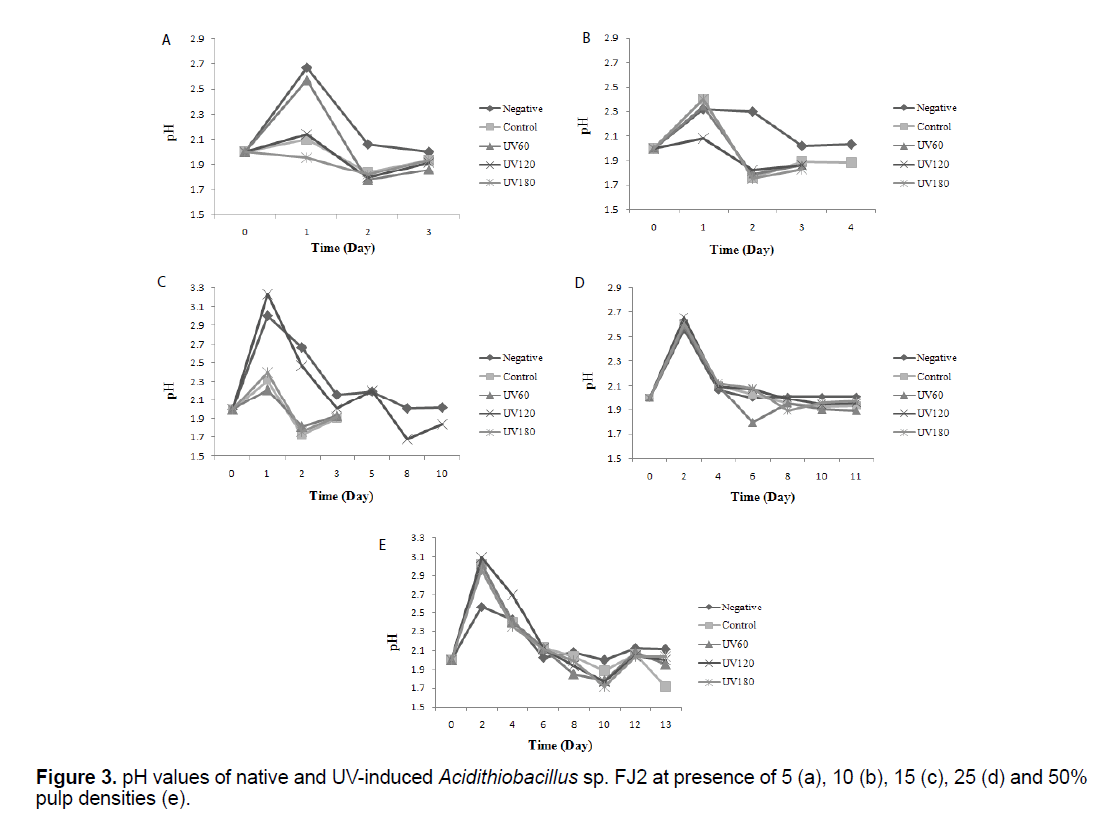
3.3 pH profiles in the uranium bioleaching processes
Figure 3 shows the results of bacterial pH variations before and after mutations at variable pulp densities. The pH values in the Figure 3, correspond to the measurement of pH before adjusting it in the optimal range of 2 for evaluating the bacterial activity. It can be seen that, for all the experiments except negative, a similar pattern was observed in which after a temporary increase in the primary days, pH values was gradually decreased to about lower 2 (p<0.05).
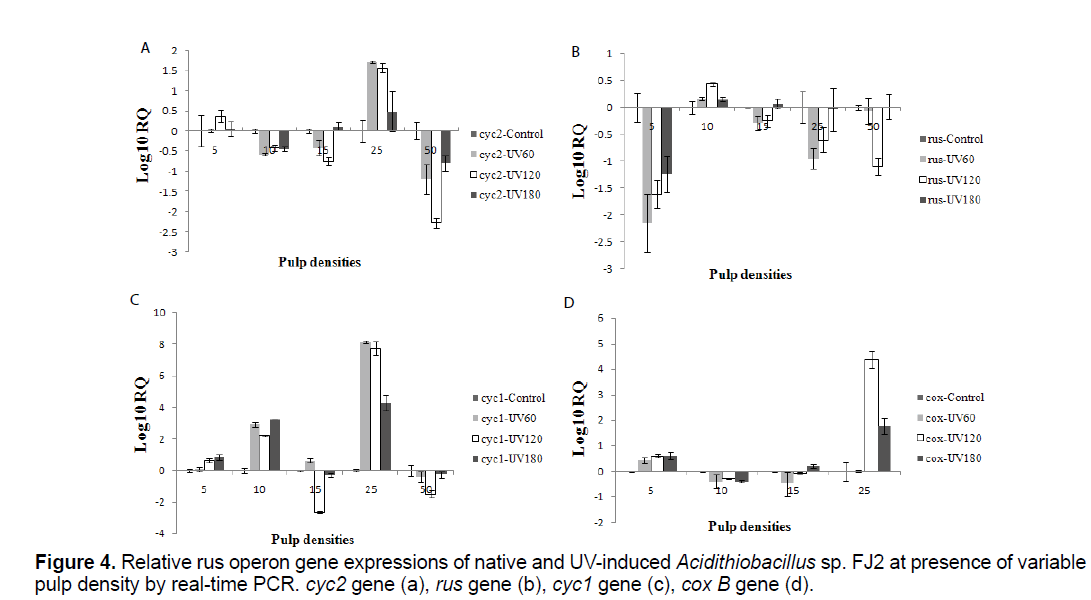
3.4 Expression of the rus operon genes at control and induced bacteria in presence of variable pulp densities
To determine the relative expressions of the rus operon genes in the presence of different concentrations of soluble uranium, control and induced bacteria were used as inocula at variable pulp densities (5%, 10%, 15%, 25% and 50%). The samples were removed at the end of bioleaching process and the cells were pelleted. Relative expression analysis of the rus operon genes was monitored by real-time PCR method (Figure 4).
The expression of rus operon genes at different situations was shown in (Figures 4a-d). Expression of these genes were seen in all the samples. In addition, the relative expressions were different at variable pulp densities. In the presence of 10% pulp density, using induced bacteria, 100% of uranium was extracted 12 hour faster than control sample. Interestingly, rus gene expression of the induced bacteria was higher than control sample in this case (p<0.05). Besides, the UV120 sample has 8 days delay in uranium extraction while, the lowest level expression of rus gene was observed in comparison with other samples at 15% pulp density (p<0.05).
In all mutants' samples, cyc2, cyc1 and cox B genes were expressed rather than the control sample at 25% pulp density (p<0.05). On the other hand, this genes expression of mutants' samples was lower than the control sample at 50% pulp density (p<0.05). In fact, cyc2, cyc1 and cox B genes of all samples approximately show a similar expression pattern at variable pulp density. Meanwhile, about cox B gene, no expression was detected in control sample at 50% pulp density. Furthermore, UV 120 sample had the lowest level expression of rus gene and amount of uranium extraction in the same pulp density.
It should be explained that, the rus expression patterns differentiated slightly. That means, the gene expression in all pulp densities and induced samples was less than control sample except 10% pulp density (p<0.05). In presence of 10% pulp density, the rus gene expression of induced bacteria was rather than the control sample (p<0.05).
4. Discussion
In this research, study the effects of UV-mutations concomitant with different pulp density considerations on relative expression of rus operon genes using native isolated Acidithiobacillus sp. FJ2 would be a novel applicable parameter in the adapted bioleaching process that would be especially unique with the achievement of the uranium dissolution at 50% pulp density with native and UV180s treated bacteria (Figure 1). These results are confirmed by Yingbo et al. [12] indicating that the UV-mutation is effective on the bioleaching process.
Our results indicated that the best levels of the bioleaching yields were achieved in low pulp densities in all control & treated samples (Figure 1). In other words, in low pulp densities, the 100% uranium yields was achieved in shorter time than in higher ore concentration (Figure 1). In this case, an increase in the rubbing between particles together with keeping away the adhesion between the particle and bacteria [16], can explain the bioleaching rate reduction in high pulp densities. Also, this rubbing may therefore cause some mechanical damage to the cell [17]. Alternatively, the toxicity from the high released doses of metals or the deteriorated environmental conditions which affected the growth and activity of the bacteria, were often the main reasons for the bioleaching efficiency declining at high pulp densities [18]. Besides that, the high pulp density resulted into high pH value of bioleaching medium. Then, the high pH value led to low activity of the bacteria resulted in low leaching efficiency of uranium (Figure 1). As known, the extraction of uranium was due to the acidic dissolution by biogenetic sulfuric acid which was generally driven by the sulfur-oxidizing bacteria [18].
On the other hand, the uranium extraction yield which would be 100% at 5%, 10%, 15% and 25%, decreased to 96.34% at 50% pulp density using UV180 s sample. In generally, the results indicated that the activities of the induced bacteria increased leading to the increased leaching rates as compared with that of the control type. This shows that UV irradiation may lead to mutations in the bioleaching bacteria which can improve the bacterial activities. The UV-induced mutagenesis is a commonly-used and useful method for bacterial breeding. The pyrimidine bases have powerful UV absorption. When UV ray is absorbed by them, the neighboring double thymines in the chain of DNA will form thymine dimer that mostly causes mutations [11]. Doudney [19] reported that UV-induced mutation depends on two radiation effects: (1) blockage of DNA synthesis by UV, necessitating the “transfer of information” through an “RNA-protein intermediate” and (2) the establishment of the photochemical modification which results eventually in mutation.
As can be seen in Figure 2, the soluble ferrous ion was decreased, while the redox potential gradually increased in all the samples. The ferrous iron oxidation which oxidize the Fe2+ to Fe3+, carried out by the bacteria in the bioleaching process. The Fe3+ is the major oxidizing agent in the sulfide ores oxidative dissolution. However, this process reduces the Fe3+ to Fe2+ which explains the importance of the bacteria in the generation and regeneration of solutions with high redox potentials that leach the sulfide ores [20]. According to the above, iron plays an important role in the bioleaching processes and the ferrous/ferric ratio has a dominating effect on the solution redox potential. Therefore, it is realized that the concentrations ratio of ferric/ferrous ions is rising, which can be observed by monitoring the redox potential that increases during the bioleaching process. Figure 2, shows that the lowest values of redox potential are observed in the negative tests curves that was probably related to the oxidation of ferrous ion by oxygen present in the solution which can improve on ferric reduction as a result of ore content [21].
The pH variation during bioleaching experiments were shown in Figure 3. It can be seen that, for all the experiments, after a transient increase, pH values was gradually decreased that mainly due to the activity of iron and sulfur-oxidizing bacteria to produce sulfuric acid. After this time, pH remained stable over the next days. The availability of the ferric iron is really influenced by the pH of the bioleaching solution and is a key factor towards the formation of the jarosite and ferric iron complex inside the bioleaching solutions, as well as influencing the conversion rate of ferrous to ferric iron. Liu et al. [11] studied the effect of pH on the jarosite formation during the bioleaching process. They noted that at the pH 2.3 and over, the inhibition of the bacterial oxidation activity was intense. This fact was attributed to the formation of a layer of precipitation on the microorganisms which prevented the diffusion of protons. The researchers revealed that as the pH of the system was increased, the jarosite formation was happened. On the other hand, Elwood Madden et al. [22] revealed that the jarosite high dissolution rate was due to the high proton substitution, or the dislocations within jarosite structures caused by changing the pH and the temperature. Interestingly, in the tests containing UV180 s sample, lower pH value were observed at 50% pulp density and in 10th day, due to a higher ferric ion concentration (oxidation product), that toward generating elemental sulfur, producing accordingly sulfuric acid as the oxidative process goes on [23].
In the final stage of tests, the relative rus operon gene expressions of all samples at variable pulp density were determined (Figure 4). In the Acidithiobacillus sp. electron transfer model, the rus operon, encodes two c-type cytochromes (Cyc1 and Cyc2), rusticyanin (rus), an ORF of unknown function (orf) and an aa3- type cytochrome oxidase (coxBACD) [5]. Rus has been shown to constitute up to 5% of total soluble proteins in Fe(II)-grown and play an essential role in the electron-transfer pathway from Fe(II) to O2 [24,25]. This protein, and the other proteins that presence in electron transfer chain, have been recommended to be involved in Fe(II) oxidation [5]. We found that, the trend of rus gene was different from the other genes of the rus operon in UV-induced samples (Figure 4). One could then question whether if all four genes are in one operon why their behavior is distinct. Indeed, Appia-Ayme et al. [5] shown that the rus gene has its own promoter and could be transcribed individually from the other genes of the operon. The rus operon is transcribed from at least three promoters that included Prus upstream from rus that transcripts are targets for posttranscriptional regulation [26,18] and PI and PII upstream from cyc2 [5]. Transcription from the PI promoter seems to depend on the pH or growth phase, while transcription from PII and Prus seems to be regulated mainly by the presence of Fe (II) [27].
On the other hand, relative rus operon gene expression of Acidithiobacillus sp. FJ2 was difference at presence of native and UV-induced bacteria in variable pulp densities (Figure 4). In fact, different dose of mutation have been influential on relative expression of rus operon genes. As can be seen in Figure 4d, there is no expression of cox B gene, at 50% pulp density in control sample, while the induced bacteria had expression in the same conditions. From these results, it could be deduced that the mutation had been a positive effect on cox B expression at presence of 50% uranium ore. In addition, the amount of ore had been very effective on rus operon genes expression. According to Tamas et al. [28], heavy metal ions interfere in protein homoeostasis and protein folding affecting cell viability. Exposure of Acidithiobacillus sp. cells to U(VI) causes a response against oxidative stress to protect cellular functions and maintain thiol homoeostasis. Downregulation of the outer membrane protein (Cyc2) expression observed in the presence of uranium suggested that a change in the permeability of the outer membrane happened, which decreasing the influx of uranium ions to the cell [28]. Whereas, U(VI) in Acidithiobacillus sp. is frequently related with the cell wall, so it is possible that a change in the outer membrane permeability occurs to decrease influx of U(VI) to the cell to prevent toxicity [29].
5. Conclusion
It seems clear from the data presented in this paper that the UV-mutation could adjust the bacterial activity leading to the high uranium extraction yield. In addition, variable pulp densities of the uranium ore and Acidithiobacillus sp. FJ2 mutations were effective on the relative expression of rus operon genes in the bioleaching process. As, we did not find any correlation between the variable pulp densities and the expression levels of rus operon genes results, It can be deduced that the bacteria may try to create efficient adaptation to sustain the bioleaching yield at different pulp densities.
References
- Mukhopadhyay A, Bhadra M, Bose K. (2005). Regional adiposity, body composition and central body fat distribution of 10-16 years old Bengalee boys of Nimta, North 24 Parganas, West Bengal, India. Coll Antropol. 29: 487-492.
- Singh A, Jagdish B, Singh GJ, et al. (2003). Essential in physical education. 5th edn. Kalyani Publishers.
- Barrow HM, McGee R. (1979). A practical to measurement in physical education. Philadelphia. Lea and Febiger.
- Fryar CD, Gu Q, Ogden CL. (2012). Anthropometric reference data for children and adults: United States, 2007-2010. National Center for Health Statistics. Vital Health Stat. 11.
- Garrett HE. (2007). Statistics in psychology and education, New Delhi: 12th edn. Paragon International Publisher.
- Johnson, Nelson. (1935). Practical measurements for evaluation in physical education. Burges Publishers.
- Shukla M, Venugopal R, Mitra M. (2008). A comparative study of growth pattern and motor quality of boys of Jawahar Navodaya Vidyalaya and Kendriya Vidyalaya in Chhattisgarh, India. J Exerc Sci Physiother. 4: 63-72.
- Michael MJ, Tim O, Arthur S, et al. (2001). International standards for anthropometric assessment. Cataloguing-in-Publishers.
- PauL PK. (2013). Age related changes on growth and motor performance of 13 & 14 years boys. Int Educ E J. 2: 99-106.
- Chakrabarty S, Bharati P. (2008). Physical growth and nutritional status of the Shabar tribal adolescents of Orissa, India: A cross-sectional study. Mal J Nutr. 14: 101-112.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences