Wolbachia Endosymbiont in the Workers of European Honeybee, Apis mellifera carnica
Mahesh Pattabhiramaiah, Dorothea Brueckner, Karl Paul Witzel and M.S.Reddy
1Honeybee Research Unit,University of Bremen,FB 2,28334,Bremen,Germany
2Max Plank Institute for Evolutionary Biology,Ploen,Germany
3Centre for Apiculture studies,Dept. of Zoology,Bangalore University,Jnana Bharathi,Bangalore-560056,India
- Corresponding Author:
- Mahesh Pattabhiramaiah
Tel: +49(0)4221-9160-206
Fax: +49(0)4221-9160-199
E-mail: reply2mahesh@gmail.com
Abstract
Invertebrates host numerous bacteria with interactions ranging from pathogenesis to symbiosis. Wolbachia is a ubiquitous, maternally inherited α-proteobacterium that causes a wide range of reproductive abnormalities in its hosts. In this paper, using polymerase chain reaction assay based on specific amplification of the 16s rRNA Wolbachia gene fragments, we recorded 100% infection in the workers of the honeybee subspecies Apis mellifera carnica collected at the Honeybee Research unit, Bremen, Germany, confirming that they are abundant in the workers of the honeybees screened
Keywords
Apis mellifera carnica,PCR,Wolbachia,16S rRNA gene.
1. Introduction
Wolbachia is one of the most common cytoplasmically inherited microorganisms in arthropods and its effects on their hosts include altering sex ratio or inducing thelytoky [1-4]. Wolbachia have been found in numerous species of Hymenoptera,including parasitoids and ants [5-8]. It can also confer fitness benefits on its host [9],and increase host resistance to infection with a range of pathogens [10-14].
At present the most common symbiotic microbe found to induce parthenogenesis is the bacterium Wolbachia pipientis. It was first identified as the microbe inducing parthenogenesis in parasitoid wasps of the genus Trichogramma [15,16]. Wolbachia induces thelytoky in at least 40 species of Hymenoptera [4]. In general Wolbachia was thought to be unique in its ability to induce parthenogenesis,but Zchori-Fein et al. [17] found an undescribed vertically transmitted bacterium also associated with parthenogenesis.
Wolbachia–host interactions are complex and range from mutualistic to pathogenic,depending on the combination of host and Wolbachia species involved. Wolbachia alter host reproduction in order to enhance the transmission of this maternally inherited agent. The presence of Wolbachia in social Hymenoptera,including the Cape bee,was investigated by Wenseleers and Billen [18] using a standard PCR protocol. Jeyaprakash et al. [19] revealed the presence of Wolbachia in Apis mellifera capensis,Apis mellifera scutellata,and their hybrid in Southern Africa.
Wolbachia infection is extremely widespread,having been found to infect 20%–75% of invertebrate species sampled [8]. Although the diversity of bacteria in the honeybee spp. Apis mellifera capensis and Apis mellifera scutellata have been well investigated [19] in African honeybee,there is a lack of such investigation in the subspecies of European honeybee,Apis mellifera carnica of Germany.
The main objective was to screen for the presence of non pathogenic endosymbiotic bacteria Wolbachia in the workers of European honeybee sub species,Apis mellifera carnica collected from honeybee research unit,University of Bremen,Germany,using a culture-independent molecular approach.
2. Materials and Methods
2.1 Colony sources
Workers of the honeybee subspecies Apis mellifera carnica was collected from different colonies of Honeybee Research Unit,University of Bremen,Germany. The samples were collected in 95% ethanol and stored at –80° C in deep freezer prior to DNA extraction.
2.2 Dissection of bees
The intestine consisting of midgut (ventriculus) and hindgut (rectum) was isolated on ice from frozen bees by clipping the stinger and the posterior segment of the abdomen with sterile forceps and carefully removing the intestine. Entire intestinal contents of adult honeybees were removed under sterile conditions,immediately frozen in liquid nitrogen,and stored at –80° C.
2.3 DNA extraction and PCR protocols
Total genomic DNA was extracted from the intestine of individual worker using Aquapure Genomic DNA kit (catalog number 7326343) reagents following the procedure suggested by the manufacturer and the genomic DNA was resuspended in 50 μl of sterile water. For PCR amplification 1 μl of genomic DNA was used for PCR amplification.
Wolbachia specific primers WOB76f and WOB1012r (Table 1),were used to amplify the Wolbachia present in the total genomic DNA,extracted from each of workers honeybee.
Standard PCR was performed by a hot start method in a 25 μl reaction volume containing 1 μl DNA sample,1 μl forward and reverse primers,5 μl 10X Buffer containing 15 mM of MgCl2,1 unit of Taq polymerase (Roche),and 1 μl dNTPs (10mM). Deionized MilliQ water was added to a final volume of 25 μl. The PCR reaction mix was prepared in one batch and then added to each sample. A sample containing deionized water in place of template DNA was included in all reactions as a negative control. PCR amplification was done on a Master Cycler Gradient (Eppendorf) under the following thermal profile: 95° C for 2min @ 1 cycle,95° C for 30sec @ 30 cycles,40°C for 30sec @ 30 cycles,73° C for 3min @ 30cycles and an extension cycle of 73° C for 1min @ 1cycle.
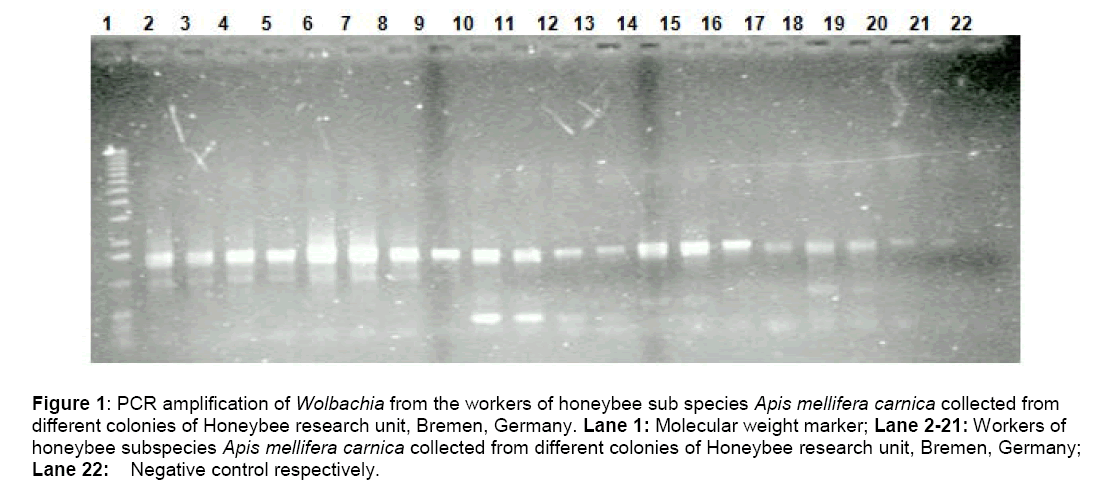
The amplified products were detected by running a 1.5% agarose gel (TAE buffer) with a 1kb molecular weight marker .The gels were stained in ethidium bromide,observed under UV Transilluminator and then photographed (Figure. 1).
Figure 1: PCR amplification of Wolbachia from the workers of honeybee sub species Apis mellifera carnica collected from different colonies of Honeybee research unit, Bremen, Germany. Lane 1: Molecular weight marker; Lane 2-21: Workers of honeybee subspecies Apis mellifera carnica collected from different colonies of Honeybee research unit, Bremen, Germany; Lane 22: Negative control respectively.
Clean laboratory practices,sealed pipette tips,and fresh reagents were used to avoid contamination. Negative controls (consisting of all components except the DNA template) were conducted on each date to detect potential contamination,but positive controls were not carried out to reduce the likelihood of contamination.
2.4 DNA sequencing and Sequence deposition
The present study was designed to develop a faster protocol for PCR amplification and sequencing of 16S rRNA gene in order to facilitate systematic studies. Specificity of the amplicons was further verified by sequencing of random samples. For this purpose,attempts were made to amplify the 16S rRNA gene by direct PCR and to sequence the amplified double-stranded DNA directly using an automated fluorescent DNA sequencer. Sequencing was carried out at the Max Plank Institute for Evolutionary Biology,Ploen,Germany. The ~1500 bp DNA samples from amplified PCR products were purified with a Prep-A-Gene PCR Clean-Up System kit (Bio-Rad) according to the manufacturer's instructions. A 500-ng amount of template (amplification product) was combined with 10 ng of primer,2 μl of Sequence buffer,and water to 10 μl. Sequencing was carried out using Big Dye 2.0 (Applied Biosystems) end-terminal cycle sequencing,followed by separation and analysis on an Applied Biosystems 3130 DNA Analysis machine.
The nucleotide sequences of directly sequenced PCR products were deposited in GenBank under accession numbers (EF032157.1,EF032158.1) using Bankit software (https://www.ncbi.nlm.nih.gov/BankIt/ index.html). A BLASTN search was carried out to find related sequences in GenBank,especially from previous reports on bacteria associated with bees and other insects.
Sequence statistics of the sequenced nucleotide (EF032157.1,EF032158.1) were analyzed using software CLC Free Workbench,Version: 5.1.1 using fasta format. Various sequence information and the statistical table including nucleotide distribution table were analyzed (Table 2,3).

3. Results and Discussion
This paper first reports the presence of Wolbachia in the workers of honeybee sub species Apis mellifera carnica collected from different colonies of Honeybee research unit,Bremen,Germany based on PCR and direct sequencing approach.
A PCR assay based on 16S primers was designed for the detection of Wolbachia pipientis in honeybee tissue,yielded an amplicon of ~900 bp,which corresponds to 16s rDNA gene of Wolbachia. All the worker honeybees were tested positive (100%) (Figure 1) for Wolbachia infection examined by the PCR. The data strongly suggest that the organism has undergone a recent and extensive spread through host populations.
So far,most studies on honeybee microflora have focused on disease-causing microorganisms [20],while much less emphasis has been given to non-pathogenic microorganisms and their impact on hosts. The inability to grow these bacteria on defined cell-free medium has been the major factor underlying these uncertainties. We circumvented this problem by selective PCR amplification and subsequent direct sequencing of the symbiont 16S rRNA genes directly from infected insect tissue.
Apart from PCR results,the BLAST analysis of the partial sequences (EF032157.1,EF032158.1) obtained from direct sequencing of random samples,revealed that the sequences were 99% identical with each other,belonging to Wolbachia pipientis.
The BLAST result of the individual sequence (Accession number EF032157),showed 99% identity with Wolbachia pipientis strain from Apis mellifera carnica (Accession number EF032158),99% identity with Wolbachia endosymbiont of Varroa destructor of Apis mellifera carnica (Accession number EF032159),99% identity with Wolbachia pipientis strain from A.diaspidis (Accession number X87407),identity of 98% with Wolbachia pipientis strain EW-p (Accession number EU096232) and 98% identity with Wolbachia W.S.P gene(Accession Z289831) ,with total query coverage of 100% and E-Value of 0.0 .
The BLAST result of the individual sequence (Accession number EF032158) showed 99% identity with Wolbachia pipientis strain from Apis mellifera carnica (Accession number EF032157),99% identity with Wolbachia endosymbiont of Varroa destructor of Apis mellifera carnica (Accession number EF032159),identity of 98% with Wolbachia pipientis strain trk1/dsz (Accession number AJ306310,AJ306309,AJ306308),and 99% identity with Wolbachia W.S.P gene(Accession Z28983),with total query coverage of 100% and E-Value of 0.0 .
The BLAST results of the partial sequence (EF032157.1,EF032158.1) verified the presence of same species of non-pathogenic Wolbachia pipientis belonging to α-proteobacteria in the worker of the honeybee subspecies Apis mellifera carnica. These bacteria may thus be specialized inhabitants of the honeybees. Therefore,these bacteria seem to constitute a specialized endogenous community in the honeybee’s alimentary tract rather than bacteria accidentally taken up from the environment passing the gut. This obligatory symbiotic relationship is evident from several reports of Jeyaprakash et al. [19],who established the presence of Wolbachia in honeybee species,Apis mellifera capensis and Apis mellifera scutellata. Interestingly the sequences (EF032157.1,EF032158.1) were 99% identical to the sequence of Wolbachia from Varroa destructor (EF032159.1) which happens to be an ectoparasite of the honeybee sub species Apis mellifera carnica,revealing the frequent horizontal transmission between the host and parasite is exclusive.
The detailed nucleotide sequence analysis of sequence (Accession no: EF032157) revealed that the length of the nucleotide was 612 bp with a molecular weight of 198,855 kDa (Table 2).The A+T content were higher than the G+C content (Table 2) with a single (N) nucleotide. It revealed considerably higher A+T content of 53.2% as compared to G+C content of only 46.5%,revealing that the sequence was AT rich.
The detailed nucleotide sequence analysis of sequence (Accession no: EF032158) revealed that the length of the nucleotide was 600 bp with a molecular weight of 195,068 kDa (Table-3).The A+T content were higher than the G+C content (Table 2) with three (N) nucleotides. It revealed a higher percentage of A+T (53%) base composition as compared to G+C content of only 45.58%,,revealing that the sequence was AT rich.
The differential sequence analysis between the Length and weight of the nucleotide sequences (Accession no: EF032157,Accession no: EF032158) was found to be 12 bp and 3.787 kDa respectively.
The A+T content of the sequence (Accession no: EF032157) was higher than the A+T content of the sequence (Accession no: EF032158) by 0.2 %. The G+C content of the sequence (Accession no: EF032157) was higher than the G+C content of the sequence (Accession no: EF032158) by 0.92%.
In insects,the relationship between the presence of Wolbachia endosymbionts and reproductive alteration of their host is well established,although the exact mechanism involved has not yet been determined. Certain Wolbachia strains have been shown to induce parthenogenesis in several parasitic wasps [3,5]. Although,Wenseleers and Billen [18] failed to demonstrate the presence of Wolbachia in Apis mellifera capensis,Jeyaprakash et al. [19] did detect Wolbachia in this subspecies. However,the same Wolbachia strain was also found in arrhenotokous honeybee workers of another subspecies,Apis mellifera scutellata,and so is unlikely to be responsible for thelytoky. Nevertheless,Jeyapraksh et al. [19] did not rule out a role for bacteria in inducing thelytoky in Cape honeybees,since other microorganisms have been shown to induce thelytoky [17,21].
The presence of Wolbachia in Apis mellifera carnica indicates a long-term and stable association between the two and may increase fecundity or have no obvious effect at all. Currently,there is no evidence that these Wolbachia strains are responsible for inducing parthenogenesis within the Hymenoptera [18],and additional work will be required to determine if these Wolbachia have a role in reproductive alterations in Apis species.
4. Conclusions
A conclusion that can be drawn from our survey is that Wolbachia spp. is highly prevalent in the workers of honeybee sub species Apis mellifera carnica in Bremen,Germany. Every colony of the worker honeybee was tested positive for Wolbachia spp. infection. This leads us to believe that Wolbachia spp. may be endemic to Apis mellifera in this area. Studies thus far indicate that Wolbachia’s effect and infection rate is not the same in every species. The amount of research on Wolbachia spp. prevalence in the Apis species of the order Hymenoptera is very limited. Further studies are needed to tell what role Wolbachia spp. may play in manipulating the reproductive abilities of Apis mellifera and also to investigate the prevalence rates of Wolbachia spp. in other honeybee species/races.
Acknowledgements
The authors are thankful to the Director,International Office,University of Bremen for financial assistance. The authors gratefully acknowledge and place in record the support and Infrastructural facilities extended by the Director,Max Plank Research Institute for Evolutionary Ecology,Ploen,Germany.
References
- Rigaud T.,Rouset F (1996). What generates the diversity of Wolbachia-arthropod interaction? Biodiv. Conserv. 5,999-1013.
- O'Neill S.L.,Hoffmann A.A.,Werren J.H (eds) (1997). Influential Passengers: Inherited Microorganisms and Arthropod Reproduction. Oxford University Press: New York,214
- Werren J.H (1997). Biology of Wolbachia. Annu Rev Entomol 42: 587–609.
- Cook J.M.,Butcher R.D.J. (1999). The transmission and effects of Wolbachia bacteria in parasitoids. Res. Popul. Ecol. 41:15-28.
- Stouthamer R (1997). Wolbachia-induced parthenogenesis,in: O'Neill S.L.,Hoffmann A.A.,Werren J.H. (Eds.),Influential passengers,inherited microorganisms and arthropod reproduction. Oxford Univ. Press,102–124.
- Shoemaker D.D.,Ross K.G.,Keller L.,Vargo E.L.,Werren J.H (2000) Wolbachia infections in native and introduced populations of fireants (Solenopsis spp.). Insect Molecular Biology,9,661– 673.
- Wenseleers T.,Ito F.,Van Borm S.,Huybrechts R.,Volckaert F.,Billen J. (1998). Widespread occurrence of the micro-organism Wolbachia in ants,Proc. R. Soc. London B 265,1447–1452.
- Jeyaprakash A.,Hoy M.A. (2000). Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of 63 arthropod species,Insect Mol. Biol. 9,393–405.
- Brownlie J.C.,Cass B.N.,Riegler M.,Witsenburg J.J.,Iturbe-Ormaetxe I.,McGraw E. A.,O’Neill S.L (2009). Evidence for metabolic provisioning by a common invertebrate endosymbiont,Wolbachia pipientis,during periods of nutritional stress. PLoS Pathog 5: e1000368.
- Teixeira L.,Ferreira A.,Ashburner M.(2008). The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biology,6,2753–2763.
- Hedges L.M.,Brownlie J.C.,O’Neill S.L.,Johnson K.N (2008). Wolbachia and virus protection in insects. Science 322: 702.
- Moreira L.A.,Iturbe-Ormaetxe I.,Jeffery J.A.,Lu G.J.,Pyke A.T.,Hedges L.M.,Rocha B.C.,Hall-Mendelin S.,Day A.,Riegler M.,Hugo L.E.,Johnson K.N.,Kay B.H.,McGraw E.A.,van den Hurk A.F.,Ryan P.A.,O'Neill S.L.,( 2009). A Wolbachia symbiont in Aedes aegypti limits infection with Dengue,Chikungunya,and Plasmodium. Cell,139,1268–1278.
- Kambris Z.,Blagborough A.M.,Pinto S.B.,Blagrove M.S.,Godfray H.C.,Sinden R.E.,Sinkins S.P(2010). Wolbachia stimulates immune gene expression and inhibits plasmodium development in Anopheles gambiae. PLoS Pathog,6: e1001143.
- Bian G.,Xu Y.,Lu P.,Xie Y.,Xi Z (2010). The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog,6: e1000833.
- Rousset F.,Bouchon D.,Pintureau B.,Juchault P.,Solignac M. (1992). Wolbachia endosymbionts responsible for various alterations of sexuality in arthropods. Proc. R. Soc. Lond. Ser. B 250:91-98.
- Stouthamer R.,Breeuwer J.A.J.,Luck R.F.,Werren H. (1993): Molecular identification of microorganisms associated with parthenogenesis. Nature 361: 66–68.
- Zchori-Fein E.,Gottlieb Y.,Kelly S.E.,Brown J.K.,Wilson J.M.,Karr T.L.,Hunter M.S. (2001). A newly discovered bacterium associated with parthenogenesis and a change in host selection behavior in parasitoid wasps. Proc Natl Acad Sci U S A 98,12555–12560.
- Wenseleers T.,Billen J. (2000) No evidence for Wolbachia-induced parthenogenesis in the social Hymenoptera. J. Evol. Biol. 13,277–280.
- Jeyaprakash A.,Hoy M.A.,Allsopp M.H (2003) Bacterial diversity in worker adults of Apis mellifera capensis and Apis mellifera scutellata (Insecta: Hymenoptera) assessed using 16S rRNA sequences. J Invertebr Pathol 84: 96-103.
- Alippi A.M.,Lopez A.C.,Aguilar O.M (2002) Differentiation of Paenibacillus larvae subsp. larvae,the cause of American foulbrood of honeybees,by using PCR and Restriction Fragment Analysis of genes encoding 16s R-rna. Appl Environ Microbiol 68: 3655-3660.
- Weeks A.R.,Reynolds K.T.,Hoffman A.A.(2002). Wolbachia dynamics and host effects: what has (and has not) been demonstrated? Trends in Ecology and Evolution 17(6): 257-262.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences