Proteome Alterations Associated With the V144D SPTLC1 Mutation That Causes Hereditary Sensory Neuropathy-I
Scott E. Stimpson, Jens R. Coorssen*, Simon J. Myers*
1Neuro-Cell Biology Laboratory,Western Sydney University,Australia;
2Molecular Physiology,Western Sydney University,Australia;
3Molecular Medicine Research Group,Western Sydney University,Australia;
4School of Science and Health,Western Sydney University,Australia;
5School of Medicine,Western Sydney University,Australia.
Received: November 16,2015; Accepted: December 11,2015; Published: December 17,2015
- Corresponding Authors:
- Jens R. Coorssen
Tel: 61 02 4620 3383
E-mail: j.coorssen@uws.edu.au - Simon J. Myers
Tel: 61 4620 3802
E-mail: s.myers@uws.edu.au
Citation: Stimpson SE,Coorssen JR,Myers SJ,Proteome Alterations Associated With the V144D SPTLC1 Mutation That Causes Hereditary Sensory Neuropathy-I. Electronic J Biol,11:4
Abstract
Background: Hereditary sensory neuropathy type I is the most common subtype and presents with clinical onset in the second to third decade of life with progressive degeneration of the dorsal root ganglion neurons. Three different missense mutations in the gene encoding for serine palmitoyltransferase long chain subunit 1 have been linked to HSN-I. Here we quantitatively assess the proteomes and identify marked protein alterations in both mitochondria and endoplasmic reticulum from HSN-I patient lymphoblasts which harbour the V144D mutation.
Methods: Mitochondria and endoplasmic reticulum were fractionated and lysed from control and patient-derived lymphoblasts. Protein samples were separated into total soluble and total membrane fractions and analysed using a well-established topdown proteomic protocol. Altered protein species were identified by LC MS/MS.
Results: Using a detailed proteomic approach, we identified 36 proteins that were completely altered in abundance in cells harbouring the V144D SPTLC1 mutation relative to normal controls.
Conclusion: The data establish that major protein alterations occur in both the endoplasmic reticulum, where the SPTLC1 protein resides, and in the mitochondria from V144D patient lymphoblasts. These proteins potentially play a major role in disease pathogenesis and may thus help to further elucidate the molecular mechanism(s) underlying hereditary sensory neuropathy type I and might also prove to be potential therapeutic targets.
Keywords
Mitochondria; Endoplasmic reticulum; SPTLC1; HSN-I; Proteomics.
Introduction
Hereditary sensory neuropathy type I (HSN-I) is the most common subtype of the HSNs [1],characterised by the progressive degeneration of the dorsal root ganglion (DRG). Onset of clinical symptoms is between the second and third decade of life [2]. Heterozygous mutations in the serine palmitoyltransferase (SPT) long chain subunit 1 (SPTLC1) have been identified as the cause of HSN-I [3,4]. The associated mutations in this gene occur at single amino acids which are highly conserved throughout different species and are therefore likely to interfere with SPT functionality and structure [5].
SPT is a pyridoxal 5'- phosphate dependent multimeric enzyme that catalyses the first step in the biosynthesis of sphingolipids,ceramide and sphingomyelin [6]. Mutations in the SPT subunits thus result in potential dysfunction and perturbations in sphingolipid synthesis and metabolism linked to a variety of diseases,in particular HSN-I [7]. As the rate determining enzyme in the de novo sphingolipid synthesis pathway,SPT is therefore a key enzyme in the regulation of cellular sphingolipid content by condensation of palmitoyl coenzyme A (CoA) with L-serine to form 3-ketodihydrosphingosine [8-10].
We recently noted altered protein expression in the mitochondria and ER from HSN-I (SPTLC1 V144D) mutant lymphoblasts [11,12]. In order to improve characterisation,a more detailed (i.e. ‘deeper’) top down analysis of the total membrane and total soluble proteomes from the mitochondria and ER of control and HSN-I (SPTLC1 V144D) patient lymphoblasts was carried out.
Numerous protein species were found to change markedly in abundance in the mitochondria and ER from the HSN-I (SPTLC1 V144D) patient lymphoblasts; these proteins are involved in energy metabolism,catalytic activity,protein transport,oxidative stress and the cytoskeleton. These protein alterations reflect the changing cellular events that underlie HSN-I.
Results
Gel images of mitochondrial and ER membrane and soluble proteins from control and patient derived lymphoblasts
All membrane and soluble protein samples were well-resolved proteomes covering the entire MW and pI range of the gels. The total numbers of resolved protein species for mitochondrial and ER membrane soluble samples are summarised in Table 1.
| Membrane | Soluble | |||
|---|---|---|---|---|
| Organelle | Control | V144D | Control | V144D |
| Mitochondrial | 550 ± 9 | 561 ± 9 | 576 ± 6 | 562 ± 7 |
| Endoplasmic Reticulum | 558 ± 6 | 577 ± 5 | 623 ± 4 | 627 ± 4 |
Table 1: Total protein species resolved by 2D from mitochondrial and ER membrane and soluble fractions obtained from controls and HSN-I (SPTLC1 V144D) patient lymphoblast.
Analysis of the membrane and soluble mitochondrial and ER protein profiles from control and HSN-I (SPTLC1 V144D) patient lymphoblasts revealed protein species alterations,change in abundance greater than or equal to 2.0 fold,in the V144D cells relative to control lymphoblasts. The analysis revealed 36 protein species that were located at varying pI / MW (kDa) coordinates. These proteins were excised; digested and LC/MS/MS analysis was carried out to identify these proteins (protein identifications are summarized in Tables 2-5).
| Spot Number | Protein Identified | Accession Number | Unique peptides matched | Sequence Coverage | Mascot Protein Score | Predicted pI | Predicted Mw (kDa) | Mascot pI | Mascot Mw (kDa) | Fold Change in V144D |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Succinate Dehydrogenase Flavoprotein Subunit, Mitochondrial | P31040 | 23 | 24% | 617 | 6.5 | 70 | 7.06 | 73.7 | 6.1 Fold á |
| 2 | Aldehyde Dehydrogenase X, Mitochondrial | P30837 | 20 | 26% | 340 | 6.1 | 50 | 6.36 | 57.6 | 2.2 Fold á |
| 3 | Calcium Binding Mitochondrial Carrier Protein SCaMc-1 | Q6NUK1 | 19 | 22% | 98 | 5.7 | 45 | 6.22 | 53.5 | 2.6 Fold â |
| 4 | Cytochrome B-C1 Complex Subunit 1, Mitochondrial | P31930 | 28 | 38% | 971 | 5.5 | 50 | 5.94 | 53.3 | 4.0 Fold á |
| 5 | Pyruvate Dehydrogenase E1 Component Subunit Alpha, Somatic Form, Mitochondrial | P08559 | 7 | 13% | 23 | 7.9 | 38 | 8.17 | 44 | 3.2 Fold á |
| 6 | Voltage-Dependent Anion-Selective Channel Protein 1 | P45880 | 10 | 26% | 265 | 8.2 | 30 | 8.62 | 31 | 2.1 Fold â |
| 7 | Ig Kappa Chain C | P01834 | 18 | 70% | 802 | 8.3 | 22 | 5.58 | 12 | Absent in V144D |
| 8 | Ig Kappa Chain C | P01834 | 15 | 63% | 704 | 6.6 | 22 | 5.58 | 12 | Only present in V144D |
Table 2: Summary table of mascot protein identification. LC-MS/MS and Mascot Database searching identified a number of proteins from control and V144D lymphoblasts isolated mitochondrial membrane proteins.
| Spot Number | Protein Identified | Accession Number | Unique peptides matched | Sequence Coverage | Mascot Protein Score | Predicted pI | Predicted Mw (kDa) | Mascot pI | Mascot Mw (kDa) | Fold Change in V144D |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Ezrin | P15311 | 55 | 55% | 1021 | 5.5 | 67 | 5.94 | 69.5 | 3.9 Fold â |
| 2 | Prolyl 4-hydroxylase subunit Alpha 1 | P13674 | 40 | 58% | 1188 | 5.4 | 60 | 5.70 | 61.3 | 3.0 Fold á |
| 3 | 60 kDa heat shock protein, Mitochondrial | P10809 | 26 | 31% | 694 | 5.2 | 58 | 5.70 | 61.2 | 2.7 Fold á |
| 4 | Dipeptidyl Peptidase 1 | Q9Y2B0 | 14 | 16% | 534 | 7.9 | 43 | 8.35 | 44 | 2.6 Fold á |
| 5 | Pyruvate dehydrogenase E1 component subunit alpha, somatic form, Mitochondrial | P08559 | 26 | 38% | 634 | 6.4 | 49 | 6.54 | 52.6 | 2.5 Fold á |
| 6 | Inorganic Pyrophosphatase 2, Mitochondrial | Q9H2U2 | 36 | 59% | 1265 | 7.0 | 32 | 7.07 | 38.4 | 2.2 Fold á |
| 7 | Pro-Cathepsin H | P09668 | 21 | 48% | 581 | 6.1 | 30 | 8.35 | 38 | 2.2 Fold á |
| 8 | Peroxiredoxin-4 | Q13162 | 36 | 69% | 1350 | 5.6 | 25 | 5.86 | 30.7 | 2.1 Fold á |
| 9 | Ig Kappa Chain C | P01834 | 31 | 88% | 1354 | 8.3 | 22 | 5.58 | 12 | Absent V144D |
| 10 | Ig Kappa Chain C | P01834 | 27 | 76% | 998 | 6.6 | 22 | 5.58 | 12 | Only present in V144D |
Table 3: Summary table of mascot protein identification. LC-MS/MS and Mascot Database searching identified a number of proteins from control and V144D lymphoblasts isolated mitochondrial soluble proteins.
| Spot Number | Protein Identified | Accession Number | Unique peptides matched | Sequence Coverage | Mascot Protein Score | Predicted pI | Predicted Mw (kDa) | Mascot pI | Mascot Mw (kDa) |
Fold Change in V144D |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Heterogeneous nuclear ribonucleoprotein D-like | O14979 | 8 | 14% | 230 | 8.1 | 40 | 9.59 | 46.6 | 3.5 Fold â |
| 2 | Serine/Threonine-protein Phosphatase PP1-Beta Catalytic Subunit | P62140 | 11 | 40% | 418 | 5.2 | 35 | 5.84 | 38 | 2.0 Fold á |
| 3 | Apolipoprotein L2 | Q9BQE5 | 11 | 42% | 210 | 5.7 | 36 | 6.28 | 37.1 | 2.8 Fold â |
| 4 | UPF0568 Protein Cl4orf166 | Q9Y224 | 18 | 56% | 315 | 6.1 | 25 | 6.19 | 28.2 | 3.3 Fold â |
| 5 | Elongation Factor 1-Beta | P24534 | 9 | 46% | 243 | 4.1 | 24 | 4.50 | 25 | 4 Fold â |
| 6 | Serine/Arginine-rich Splicing Factor 3 | P84103 | 6 | 36% | 152 | 5.8 | 17 | 9.64 | 19.5 | 2.9 Fold â |
| 7 | Ig Kappa Chain C | P01834 | 26 | 80% | 1303 | 6.6 | 22 | 5.58 | 12 | Only present in V144D |
| 8 | Ig Kappa Chain C | P01834 | 22 | 80% | 1065 | 8.8 | 22 | 5.58 | 12 | Absent from V144D |
Table 4: Summary table of mascot protein identification. LC-MS/MS and Mascot Database searching identified a number of proteins from control and V144D lymphoblasts isolated ER membrane proteins.
| Spot Number | Protein Identified | Accession Number | Unique peptides matched | Sequence Coverage | Mascot Protein Score | Predicted pI | Predicted Mw (kDa) | Mascot pI | Mascot Mw (kDa) |
Fold Change in V144D |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Peroxiredoxin-2 | P32119 | 19 | 65% | 453 | 5.0 | 20 | 5.66 | 45.3 | 2.4 Fold á |
| 2 | Lymphocyte-Specific Protein 1 | P33241 | 24 | 49% | 901 | 3.5 | 50 | 4.69 | 37.4 | 3.2 Fold á |
| 3 | Proteasome Activator Complex Subunit 1 | Q06323 | 10 | 37% | 357 | 6.0 | 35 | 5.78 | 28.9 | 4.2 Fold á |
| 4 | Proteasome Subunit Alpha Type-3 | P25788 | 18 | 41% | 372 | 5.2 | 30 | 5.19 | 28.6 | 2.6 Fold á |
| 5 | Protein CDV3 Homolog | Q9UKY7 | 14 | 64% | 578 | 6.2 | 60 | 6.06 | 27.3 | 2.3 Fold â |
| 6 | Chloride intracellular channel protein 1 | O00299 | 38 | 68% | 1174 | 5.2 | 30 | 5.09 | 27.2 | 2.7 Fold á |
| 7 | Adenine Phosphoribosyltransferase | P07741 | 24 | 72% | 876 | 5.1 | 17 | 5.78 | 19.8 | 3.0 Fold â |
| 8 | Eukaryotic Translation Initiation Factor 5A-1 | P63241 | 26 | 70% | 898 | 4.3 | 15 | 5.08 | 17 | 2.1 Fold á |
| 9 | Ig Kappa Chain C | P01834 | 21 | 80% | 1278 | 6.6 | 22 | 5.58 | 12 | Only present in V144D |
| 10 | Ig Kappa Chain C | P01834 | 29 | 86% | 1609 | 8.3 | 22 | 5.58 | 12 | Absent in V144D |
Table 5: Summary table of mascot protein identification. LC-MS/MS and Mascot Database searching identified a number of proteins from control and V144D lymphoblasts isolated ER soluble proteins.
Functions of the identified proteins in mitochondrial and ER fractions
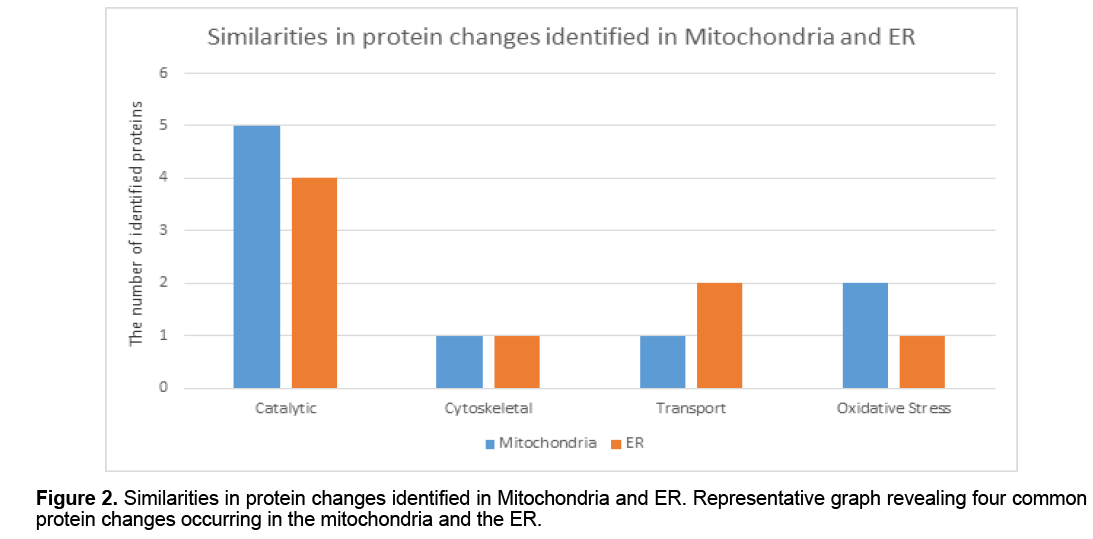
Proteins identified from mitochondrial and ER membrane and soluble fractions were grouped based upon their biological functionality; a pie graph has been used to provide a visual analysis of protein changes in the HSN-I (SPTLC1 V144D) disease state (Figure 1). This analysis indicates that the majority of the protein alterations in mitochondria are involved in catalytic activity,cytoskeleton,transport,oxidative stress,calcium binding and energy metabolism. However,whist there is some overlap in terms of alterations to mitochondrial and ER proteins (Figure 2 ),many of those identified from the ER function in the areas of protein biosynthesis,apoptosis,cell proliferation,protein binding and lipid binding (Figure 1).
Discussion
SPT is the key rate determining enzyme in sphingolipid metabolism. Mutations within the SPTLC1 subunit thus result in potential perturbations in sphingolipid synthesis and metabolism that may be the underlying causative effects of HSN-I [7]. In initial studies,we showed that a number of mitochondrial and ER proteins are altered in abundance,correlating with the SPT mutations in patient derived cells [11-13]. Here we have carried out a more detailed top-down proteomic analysis and identified 36 protein species that change in abundance in the mitochondria and ER of HSN-I patient cells.
To identify potentially critical protein changes,mitochondria and ER were first isolated from control and HSN-I (SPTLC1 V144D) patient lymphoblasts and further separated into total membrane and total soluble protein fractions prior to high resolution topdown proteomic analyses using 2DE [14-16]. These analyses revealed numerous protein changes in both the membrane and soluble protein fractions from the control and HSN-I (SPTLC1 V144D) patient lymphoblasts. Mitochondrial protein species that changed in abundance were involved in catalytic activity,cytoskeleton,protein transport,oxidative stress,calcium binding and energy metabolism (Figure 1). The proteins identified in the ER fractions were involved in catalytic activity,cytoskeleton,and lipid binding (Figure 1). While there were a number of non-related protein species that changed in the mitochondria compared to the ER,there were a number of similarities in biological processes,most notably catalytic activity,cytoskeleton,protein transport and oxidative stress [17-19] (Figure 2 ).
Mitochondria are known to play a role in neurodegeneration,and structural alterations have been characterised in V144D patient lymphoblasts [20],with further studies identifying changes at the protein level within isolated mitochondria [11]. The higher resolution analyses here provide more detailed information still. Oxidative stress can have an impact upon the cell,causing severe and extensive damage including protein aggregation and impaired ion transport [21]. Previously,ubiquinol cytochrome C subunit 1 was found to increase in abundance in V144D patient lymphoblasts [11]; here this protein was found to increase in abundance (i.e. 4-fold),and this was accompanied by a 2.1- fold increase in the abundance of Peroxiredoxin-4,a protein with antioxidant functions,that reduces the build-up of hydrogen peroxide via a thiol-dependent cycle [21]. These findings correlate with a potential increase in reactive oxygen species (ROS) within the disease cells,which could lead to further disruption to mitochondria.
Perturbations to energy production within neurons,having high metabolic demands,can have catastrophic consequences. Succinate dehydrogenase flavoprotein and pyruvate dehydrogenase E1 subunit are both part of the electron transport chain,and both are increased in abundance (i.e. 6.1 and 3.2 fold,respectively),likely highlighting an energy metabolism issue within the mitochondria [22]. In addition to a potential need to increase energy output,increased levels of succinate dehydrogenase could also potentially increase superoxide formation [22]. Whether these proteins apparently increased abundance is due to a direct need for increased energy or a compensatory effect due to an increase in ROS production and oxidative stress disrupting the electron transport chains ability to produce energy remains unclear,but a destructive spiral would seem a distinct possibility.
Ca2+ is also required for energy production within mitochondria,but increased Ca2+ levels can lead to free radical generation [23]. The data identifies a 2.6-fold decrease in the Ca2+ binding mitochondrial carrier protein (ScaMc-1). This decrease might be a protective mechanism due to the already high levels of ROS but will also cause a decrease in ATP production within the mitochondria. Voltage dependent anion selective channel protein 1 (VDAC) allows mitochondrial influx/efflux of metabolites such as ATP,and may also have a role in regulating Ca2+ in mitochondria [24]. A decrease in VDAC in the V144D mutant,in conjunction with the reduction of SCaMc-1 could result in an overall decrease in intracellular Ca2+ levels in mitochondria and thus decreased ATP production,again strengthening the possibility for a destructive circle of cellular events.
Dipeptidyl peptidase 1,also known as Cathepsin C and Pro-cathepsin H has been shown to be proapoptotic by cleaving Bid and Blc-2 family proteins released by mitochondria; greatly increasing the cascade of caspase apoptotic factors to be released [25,26]. The abundance of these proteins are increased in the mutant cells indicating a link to increased mitochondrial apoptotic process occurring (2.6 and 2.2 fold increase respectively).
Eukaryotic translation initiation factor 5A-1 (eIF5A) has been shown to regulate the Bcl-2 binding protein P53 and the P53 apoptosis pathway. In addition,eIF5A has a regulatory function in protein synthesis. Its increase in abundance of 2.1 fold in the V144D mutant may possibly be the cells response to stabilise uncontrolled protein misfolding due to ER stress [27]. Peroxiredoxin-2 was detected in the ER with an increase in abundance of 2.4 fold,peroxiredoxin-2 just like peroxiredoxin-4 found in the mitochondrial fraction is an antioxidant [28],potentially increased in the mutant state in response to increased ROS production occurring within the ER and throughout the cell.
ER stress may decrease mRNA to reduce the protein load upon the ER to help reduce the amount of misfolded proteins being produced [29],thus we see reduced levels of the serine/threonine phosphatase PP-1,serine/arginine-rich splicing factor and Elongation factor 1-beta within HSN-I (SPTLC1 V144D) patient lymphoblasts. With known ER stress occurring [20] the reduction in abundance of these proteins could be as a result of a compensatory effect reducing the load of protein synthesis occurring in the stressed ER.
Lymphocyte specific protein 1 is an F-actin binding protein [30],it’s 3.2 fold increase in abundance,correlates with other cytoskeletal changes observed in the HSN-I (SPTLC1 V144D) patient lymphoblasts suggesting that maintenance of the cytoskeleton is being increased potentially due to increasing amount of ROS,known to cause actin remodelling and potential axonal retraction in the neuron [31].
Proteasome activator complex 1 (PSME1) and proteasome subunit alpha type 3 (PSMA3),degrade misfolded proteins,in an ubiquitin dependent process [32,33]. Both these proteins are increased in abundance,4.2 and 2.6 fold respectively in the V144D patient derived lymphoblasts. Previous studies have identified Ubiqutin-40s Ribosomal Protein S27a [12],as such we see here the increase in proteasomes correlating a potential increase in the number misfolded proteins directly due to ER stress,oxidative stress or by another mechanism that affects protein conformation.
Bcl-2 family proteins are regulators of mitochondrial derived apoptosis. Bcl-2 proteins can illicit or inhibit cell death. Apolipoprotein L2 (ApoL2) has a potential apoptotic role being a BH3- protein,localising to mitochondria. This region,known as the ‘BH3- domain’,is essential for the apoptotic function of Bcl- 2 autophagy,while the exact role of ApoL2 remains to be determined,a reduction in the V144D diseased state may cause dysregulation of authopghy [34].
Interestingly,the Chloride intracellular channel protein 1 was identified with a 2.7 fold increased abundance in the HSN-I (SPTLC1 V144D) patient lymphoblasts. We have previously reported this proteins increased expression within the HSN-I (SPTLC1 V144D) patient lymphoblasts [12]. It acts as a chloride-selective ion channel and usually exists in a soluble form in the cytoplasm and nucleoplasm [35],but following stimuli undergoes major structural changes and inserts in lipid membranes,where cell oxidation appears to be an important stimuli determining the transition of Chloride intracellular channel protein 1 between these two forms [36].
It was identified that there were four other proteins with a marked absence or presence in all mitochondrial and ER fractions. These protein species were located at 24 kDa,but each had a different pI (6.6 and 8.3 respectively). Following mass spectral analysis,these proteins identified as Ig Kappa Chain C. This finding correlates with our previous studies in the HSN-I (SPTLC1 V144D) patient lymphoblasts [11,12].
Conclusion
This investigation has shown a correlation between previous studies revealing an increase in proteins induced by oxidative stress and mitochondrial electron transport chain proteins. This study also identified changes in calcium channel proteins,cytoskeletal proteins,energy transport proteins. Some of these findings reflect previous studies carried out,providing more evidence for a link of increased misfolded proteins,oxidative stress,and cytoskeleton remodelling and potential changes in Ca2+ signalling within the mitochondria. With mounting discoveries into protein alterations in the V144D mutation it may provide a greater in-sight into the molecular mechanisms that are occurring in HSN-I.
Materials
All cell culture stock solutions,including RPMI- 1640,Foetal Bovine Serum (FBS),Penicillin (100 U/ mL),Streptomycin (100 μg/mL),L-glutamine (2 M),HEPES (1 M),and phosphate buffered saline (PBS) were purchased from GIBCO Invitrogen (Australia). Cell culture consumables were purchased from BD Falcon (Greiner,USA).
Methods
EBV transformed lymphoblasts
EBV transformed control and V144D HSN-I patient lymphoblasts were kindly provided by Prof. Garth Nicholson (Molecular Medicine Laboratory,Anzac Research Institute,Sydney) [13].
Lymphoblast cultures
Lymphoblasts were cultured in RPMI-1640 media (GIBCO),supplemented with FBS (10% v/v),Penicillin (1 U/mL),Streptomycin (1 μg/mL),L-glutamine (2 mM),and HEPES (1 mM) at 37oC in a humidified atmosphere of 5% CO2,using T75 cm2 culture flasks (Greiner,Interpath). Prior to use in biochemical assays,lymphoblasts were collected by centrifugation at 1,500 x g (5 min at RT) and washed in PBS. Cell counts were obtained using the Countess Automated Cell Counter (Invitrogen,Australia).
Isolation of mitochondrial proteins
Briefly,mitochondria were isolated using a sucrose density gradient [14,15]. Lymphoblasts were first centrifuged at 1,500 x g for 5 min,and the cells were then washed in 10 ml of ice cold 1X PBS prior to suspension in 10 ml ice cold CaSRB Buffer (10 mM NaCl,1.5 mM CaCl,10 mM Tris-HCL,pH 7.5) and left on ice for 10 min. Cells were homogenised using a Dounce homogenizer (Kimble-Chase,USA) and 7 mL of 2.5X MS buffer (210 mM Mannitol,70 mM sucrose,5 mM EDTA,5 mM Tris-HCl,pH 7.6) was added to restore isotonicity. Homogenates were centrifuged at 700 x g for 5 min to remove nuclei and unbroken cells. The resulting supernatant was centrifuged at 15,000 x g for 10 min to pellet the crude mitochondria. Sucrose gradients were made in 4 mL high speed centrifuge tubes (Beckman Coulter,USA) by adding 1 mL of 1.7 M sucrose buffer (1.7 M sucrose,10 mM Tris-base,0.1 mM EDTA,pH 7.6) overlayed with 1.6 mL of 1.0 M sucrose buffer (1.0 M sucrose,10 mM Tris-base,0.1 mM EDTA,pH 7.6). The mitochondrial pellet was resuspended in 1.6 mL of 1x MS buffer and overlayed on top of the sucrose gradient and centrifuged at 40,000 x g for 30 min. The mitochondrial band,in the middle of the gradient,was gently removed using a 20 G needle,transferred to a 1.5 mL tube,and centrifuged at 16,000 x g for 15 min. The resulting pellet was resuspended in 2D solubilisation buffer containing 8 M urea,2 M thiourea,4% (w/v) CHAPS and a cocktail of protease inhibitors.
Isolation of ER proteins
Briefly,[14,15] Lymphoblasts were first centrifuged at 1,500 x g for 5 min,and the cells were then washed in 10 ml of ice cold 1X PBS prior to suspension in 10 mL ice cold CaSRB Buffer (10 mM NaCl,1.5 mM CaCl,10 mM Tris-HCL,pH 7.5) and left on ice for 10 min. Cells were homogenised using a Dounce homogenizer (Kimble-Chase,USA) and 7 mL of 2.5 X MS buffer (210 mM Mannitol,70 mM sucrose,5 mM EDTA,5 mM Tris-HCl,pH 7.6) was added to restore isotonicity. Homogenates were centrifuged at 700 x g for 5 min to remove nuclei and unbroken cells. The resulting supernatant was centrifuged at 15,000 x g for 10 mins to remove mitochondria. A sucrose gradient was made in 15 mL high speed centrifuge tubes (Beckman Coulter,USA) by adding 2 mL of 2.0 M sucrose buffer (2.0 M sucrose,10 mM Tris-base,0.1 mM EDTA,pH 7.6) overlayed with 3.0 mL of 1.5 M sucrose buffer (1.5 M sucrose,10 mM Tris-base,0.1 mM EDTA,pH 7.6) and 3.0 mL of 1.3 M sucrose (1.3 M sucrose,10 mM Tris-base,0.1 mM EDTA,pH 7.6) ER containing supernatant was loaded on top of the sucrose gradient and spun at 152,000 x g for 70 min. The ER band,the interface of the 1.5 M and 1.3 M sucrose,was gently removed using a 20 G needle,transferred to a 4mL high speed centrifuge tubes (Beckman Coulter,USA) and centrifuged at 100,000 x g for 35 min. The resulting pellet was resuspended in 2D solubilisation buffer containing 8 M urea,2 M thiourea,4% (w/v) CHAPS and a cocktail of protease inhibitors.
Membrane and soluble protein fractionation
Harvested mitochondrial and ER proteins were separated into membrane and soluble protein fractions as previously described [16]. Briefly; isolated proteins were placed in 20mM HEPES for 3 min on ice with an equal volume 2X PBS subsequently added. Membranes were collected at 125 000 x g for 3 h. The supernatant was collected and membrane pellet resuspended in 1X PBS and spun at 125 000 x g for a further 3 h. Washed membranes were solubilised in 2D solubilisation buffer containing 8 M urea,2 M thiourea,4% (w/v) CHAPS. Soluble protein fractions were concentrated using a 3 kDa cut-off Millipore Amicon Ultra Centrifugal filters and resuspended in 4M Urea.
Protein concentration
Determination of total cellular protein was performed using the EZQ Protein Estimation Assay (Invitrogen,Australia) as previously described [17].
Two dimensional gel electrophoresis
Protein concentration estimations (EZQ assay) were performed on patient and control mitochondrial and ER protein fractions; a total of 100 μg protein was used for each 2DE analysis. 2DE was carried out as previously described [11,16,18,19]; briefly,proteins were reduced and alkylated in solutions containing total protein extraction buffer (containing 8M urea,2M thiourea and 4% CHAPS without ampholytes),total extraction buffer with 2% ampholytes,TBP/ DTT disulphide reduction buffer (2.3 mM Tributyl phosphine and 45 mM DTT) and alkylation buffer (230 mM acrylamide monomer).
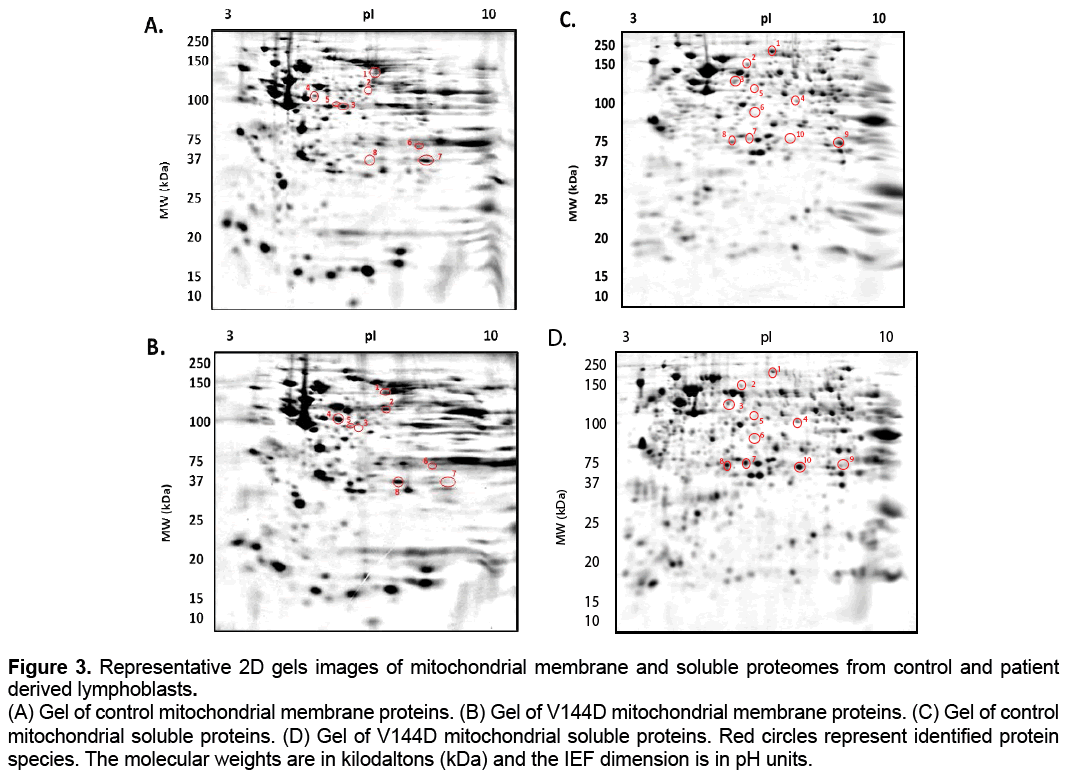
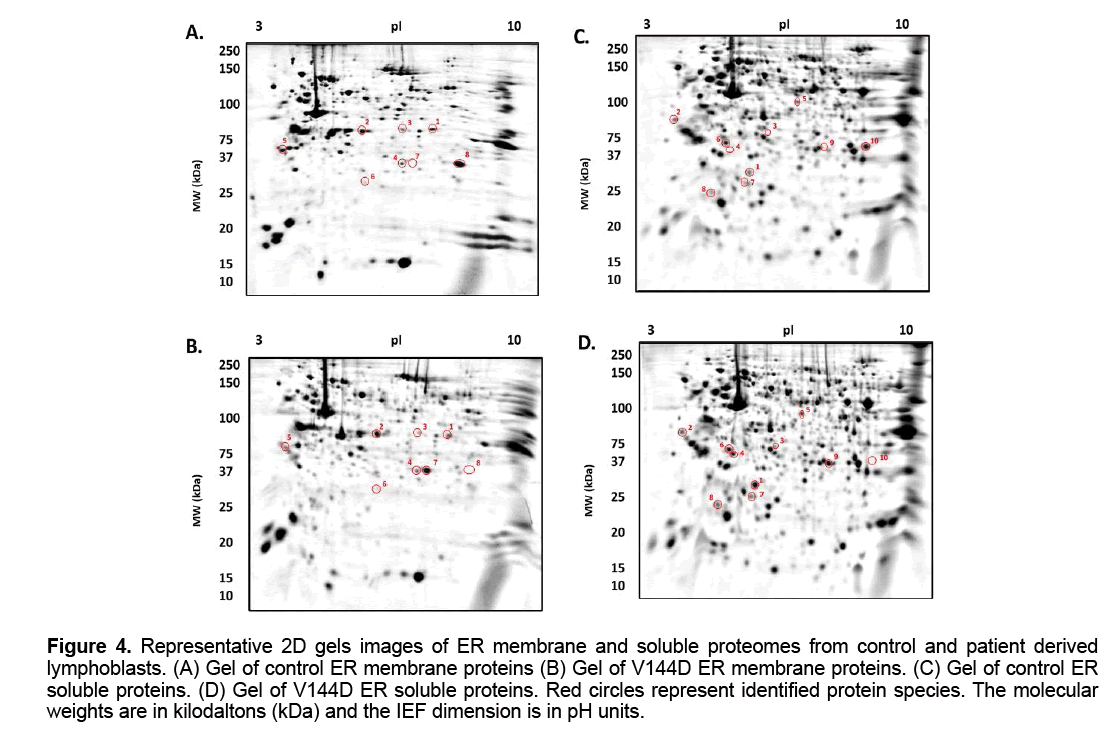
The treated samples were added to 7 cm Non- Linear pH 3-10 IPG strips (Bio-Rad ReadyStrip),and rehydrated for 16 h at RT. Isoelectric focusing (IEF) was then carried out at 20° C using the Protean IEF Cell (Bio-Rad,USA). After IEF,IPG strips were then resolved in the second dimension using a 12.5% T,2.6% C polyacrylamide gel buffered with 375 mM Tris buffer (pH 8.8),0.1% (w/v) sodium dodecyl sulphate and polymerised with 0.05% (w/v) ammonium persulphate and 0.05% (v/v) tetramethylethylenediamine (TEMED). A stacking gel containing a 5% T,2.6% C polyacrylamide buffered with 375 mM Tris buffer (pH 8.8),0.1% (w/v) SDS and included 0.1% bromophenol blue was added to the resolving gel. The IPG strips were placed onto the stacking gel and overlaid with 0.5% (w/v) low melting agarose dissolved in 375 mM Tris (pH 8.8),with 0.1% (w/v) SDS. Electrophoresis was carried out at 4° C; 150V initially for 10 min then reduced to 90V for 2.5 h. The gels were placed in fixative containing 10% methanol and 7% acetic acid for 1 h. The gels were washed with distilled water for 20 min,3 times and subsequently stained with colloidal coomassie blue (0.1% (w/v) CCB G-250,2% (v/v) phosphoric acid,10% (w/v) ammonium sulphate,20% (v/v) methanol) for 20 h,with constant shaking at RT [18]; the gels were the de-stained 5 times with 0.5 M NaCl,15 min each. Imaging of CBB-stained gels on the FLA-9000 imager (FUJIFILM,Tokyo,Japan) was carried out at 685/750 excitation/emission with a photomultiplier tube (PMT) setting of 600 V and pixel resolution set to 100 μm [18]. Analysis of 2D gel images was performed using Delta 2D software (version 4.0.8; DECODON GmbH,Gerifswald,Germany) with automated spot detection (Local Background Region: 96; Average Spot Size: 32 and sensitivity in percentage: 20.0) (Figure 3 and 4).
Figure 3: Representative 2D gels images of mitochondrial membrane and soluble proteomes from control and patient
derived lymphoblasts.
(A) Gel of control mitochondrial membrane proteins. (B) Gel of V144D mitochondrial membrane proteins. (C) Gel of control
mitochondrial soluble proteins. (D) Gel of V144D mitochondrial soluble proteins. Red circles represent identified protein
species. The molecular weights are in kilodaltons (kDa) and the IEF dimension is in pH units.
Figure 4: Representative 2D gels images of ER membrane and soluble proteomes from control and patient derived lymphoblasts. (A) Gel of control ER membrane proteins (B) Gel of V144D ER membrane proteins. (C) Gel of control ER soluble proteins. (D) Gel of V144D ER soluble proteins. Red circles represent identified protein species. The molecular weights are in kilodaltons (kDa) and the IEF dimension is in pH units.
Mass spectrometry
For analysis a selection criteria was applied. For inclusion,changes in mean normalised spot volume (the abundance of resolved protein species) had to be greater than or equal to a 2.0 fold increase or decrease between control and HSN-I (SPTLC1 V144D) patient lymphoblast,have a p-value <0.05 and be present in all replicate gels [18,19]. The protein species of interest were excised from gels and de-stained overnight. The gel pieces were then reduced and alkylated in 10 mM Dithiothreitol (DTT) and 15 mM Idoacetic acid (IAA),and subsequently incubated with trypsin solution (10 ng/μL,pH 7.4) for 16 h at 37ºC. LC-MS/MS analysis was carried out on a nanoAquity UPLC (Waters Corp.,Milford,MA,USA) linked to a Xevo QToF mass spectrometer from Waters (Micromass,UK). The data were acquired using Masslynx software (Version 4.1,Micromass UK). The MS/MS data files were searched against SwissProt databases with semi-trypsin as the enzyme. The following parameters were used in Mascot for identification of the peptides: maximum missed cleavage of 2,positive peptide charge of 2,3 and 4,peptide mass tolerant of 0.5 Da in MS and MS/ MS data base,fixed modification: carbamidomethyl (C) and variable modifications: oxidation (M).
Competing Interests
The authors have no competing interests.
Author Contributions
SES carried out all experimentation,data analysis and wrote an initial draft of the manuscript; JRC participated in design of study,provided access to the proteomics facility in which the bulk of the work was carried out,and re-drafted substantial portions of the draft; SJM conceived the study,participated in design of study,re –drafted substantial sections of the drafts. All authors read and approved the final manuscript.
Acknowledgements
We are grateful to Prof Garth Nicholson (Molecular Medicine Laboratory and Northcott Neuroscience Laboratory Anzac Research Institute,Sydney) for providing all EBV transformed lymphoblast lines used in this study. SES was supported by an APA Research Scholarship,and the UWS School of Science and Health Postgraduate research fund. SJM notes the continuing support of an anonymous Private Foundation. JRC acknowledges the support of the UWS School of Medicine.
List of Abbreviations
2DE,two dimensional gel electrophoresis; Apol2,Apolipoprotein L2.; DRG,dorsal root ganglion; DTT,Dithiothreitol; eIF2A,Eukaryotic translation factor 5A- 1; ER,endoplasmic reticulum; FBS,Foetal Bovine Serum; HSN,Hereditary sensory neuropathies; HSN-I,Hereditary sensory neuropathy type I; IAA,Idoacetic acid; IEF,Isoelectric focusing; kDa,Kilodaltons; LCB1,long-chain base one; LC/MS,liquid chromatography/ mass spectrometry; PBS,phosphate buffered saline; PMT,photomultiplier tube; PSME1,Proteasome activator complex subunit 1; PSMA3,Proteasome subunit alpha type-3; ROS,reactive oxygen species; SCaMc-1,Calcium binding mitochondrial carrier protein 1; SPT,serine palmitoyltransferase; SPTLC1,serine palmitoyltransferase long chain subunit 1; TEMED,tetramethylethylenediamine; VDAC,Voltage dependent anion-selective channel protein 1.
References
- Dyck PJ,Thomas PK.( 2005). Dyck: Peripheral Neuropathy,4th Edition. Philadelphia,Mosby Elsevier.
- VerhoevenK,Coen K,De Vriendt E,et al. (2004). SPTLC1 mutation in twin sisters with hereditary sensory neuropathy type I. Neurology. 62: 1001-2.
- Bejaoui K,Wu C,Scheffler MD,et al. (2001).SPTLC1 is mutated in hereditary sensory neuropathy,type 1. Nat Genet. 27: 261-2
- Dawkins JL,Hulme DJ,Brahmbhatt SB,Auer-Grumbach M,Nicholson G A.(2001). Mutations in SPTLC1,encoding serine palmitoyltransferase,long chain base subunit-1,cause hereditary sensory neuropathy type I. Nat Gene.27:309-12.
- Verhoeven K,Timmerman V,Mauko B,et al.(2006). Recent advances in hereditary sensory and autonomic neuropathies. CurrOpin Neurol. 19: 474-80.
- Hornemann T,Richard S,Rutti M,Wei Y,Von-Eckardstein A. (2006) ‘Cloning and initial characterization of a new subunit for mammalian serine-palmitoyltransferase’.The Journal of biological chemistry.49: 37275-37281.
- WeiJ,Yerokun Y,Liepelt M,et al. (2007). 2-1 Serine Palmitoyltransferase. Sphingolipid Biology. Springerlink. 25-27.
- Mandon EC,Ehses I,Rother J,Van Echten G,Sandhoff K. (1992). Subcellular localization and membrane topology of serine palmitoyltransferase,3-dehydrosphinganine reductase,and sphinganine N-acyltransferase in mouse liver. J Biol Chem. 267: 111-448.
- Yard B,Carter L,Johnson K,et al. (2007).The structure of serine palmitoyltransferase; gateway to sphingolipidbiosynthesis.Journal of molecular biology. 370: 870-886.
- Yasuda S,Nishijima M,Hanada K. (2003). Localization,topology,and function of the LCB1 subunit of serine palmitoyltransferase in mammalian cells. J Biol Chem. 278: 4176-83.
- StimpsonSE,Coorssen JR,Myers SJ. (2014). Mitochondrial protein alterations in a familial peripheral neuropathy caused by mutations in the sphingolipid protein,SPTLC1. J Chem Biol.
- Stimpson,SE,Coorssen JR,Myers SJ. (2015). Isolation and identification of ER associated proteins with unique expression changes specific to the V144D SPTLC1 mutations in HSN-I. BMC Neuroscience.
- Dedov V,Dedova I,Merrill A,Nicholson G. (2004).Activity of partially inhibited serine palmitoyltransferase is sufficient for normal sphingolipid metabolism and viability of HSN1 patient cells.Biochimica et biophysica acta.1688: 168-175.
- Bozidis P,Williamson CD,Colberg-Poley AM. (2007). Isolation of endoplasmic reticulum,mitochondria,and mitochondria-associated membrane fractions from transfected cells and from human cytomegalovirus-infected primary fibroblasts. Curr Protoc Cell Biol.27.
- Vaseva AV, Moll UM. (2013). Identification of p53 in mitochondria. Methods Mol Biol.962: 75-84.
- Butt RH,Coorssen JR. (2005). Postfractionation for enhanced proteomic analyses: routine electrophoretic methods increase the resolution of standard 2D-PAGE. J Proteome Res.4: 982-91.
- Churchward M,Butt RH,Lang J,Hsu K,Coorssen J. (2005). Enhanced detergent extraction for analysis of membrane proteomes by two-dimensional gel electrophoresis. Proteome Sci. 3: 5.
- Gauci VJ,Padula MP,Coorssen JR.(2013). Coomassie blue staining for high sensitivity gel-based proteomics. J Proteomics. 90: 96-106.
- Wright EP,Partridge MA,Padula MP,et al. (2014). Top-down proteomics: Enhancing 2D gel electrophoresis from tissue processing to high-sensitivity protein detection. Proteomics.14: 872-89.
- Myers S,Malladi C,Hyland R,et al. (2014). Mutantions in the SPTLC1 protein cause mitochondrial structual abnormalisites and endoplasmic reticulum stress in lymphoblasts. DNA and Cell Biology.33: 7.
- Tavender TJ,Bulleid NJ. (2010). Peroxiredoxin IV protects cells from oxidative stress by removing H2O2 produced during disulphide formation. J Cell Sci.123: 2672-9.
- Guzzo G,Sciacovelli M,Bernardi P,Rasola A. (2014). Inhibition of succinate dehydrogenase by the mitochondrial chaperone TRAP1 has anti-oxidant and anti-apoptotic effects on tumor cells. Oncotarget. 5: 11897-908.
- Feissner RF,Skalska J,Gaum WE,Sheu SS. (2009). Crosstalk signaling between mitochondrial Ca2+ and ROS.Front Biosci (Landmark Ed). 14: 1197-218.
- Brookes PS,Yoon Y,Robotham JL,Anders MW,Sheu SS. (2004). Calcium,ATP,and ROS: a mitochondrial love-hate triangle.Am J Physiol Cell Physiol. 287: C817-33.
- Droga-Mazovec G,Bojic L,Petelin A,et al. (2008). Cysteine cathepsins trigger caspase-dependent cell death through cleavage of bid and antiapoptotic Bcl-2.
- Turk V,Stoka V,Vasiljeva O,et al. (2012). Cysteine cathepsins: from structure,function and regulation to new frontiers. BiochimBiophysActa. 1824: 68-88.
- Huang Y,Higginson DS,Hester L,Park MH,Snyder SH. (2007). Neuronal growth and survival mediated by eIF5A,a polyamine-modified translation initiation factor. ProcNatlAcadSci U S A.104: 4194-9.
- Ogasawara Y,Ohminato T,Nakamura Y,Ishii K. (2012). Structural and functional analysis of native peroxiredoxin 2 in human red blood cells. Int J Biochem Cell Biol.44: 1072-7.
- Kawai T,Fan J,Mazan-Mamczarz K,Gorospe M. (2004). Global mRNA stabilization preferentially linked to translational repression during the endoplasmic reticulum stress response. Mol Cell Biol.24: 6773-87.
- Liu L,Cara DC,Kaur J,et al. (2005). LSP1 is an endothelial gatekeeper of leukocyte transendothelial migration. J Exp Med. 201: 409-18.
- Hallengren J,Chen PC,Wilson SM.(2013). Neuronal ubiquitin homeostasis. Cell BiochemBiophys.67: 67-73.
- Johnston SC,Whitby FG,Realini C,Rechsteiner M,Hill CP.(1997). The proteasome 11S regulator subunit REG alpha (PA28 alpha) is a heptamer.Protein Sci.6: 2469-73.
- Shi Z,Li Z,Li ZJ,et al. (2014). Cables1 controls p21/Cip1 protein stability by antagonizing proteasome subunit alpha type 3. Oncogene.
- Galindo-Moreno J,Iurlaro R,El Mjiyad N,et al. (2014). Apolipoprotein L2 contains a BH3-like domain but it does not behave as a BH3-only protein. Cell Death Dis. 5: e1275.
- Warton K,Tonini R,Fairlie WD,et al.(2002). Recombinant CLIC1 (NCC27) assembles in lipid bilayers via a pH-dependent two-state process to form chloride ion channels with identical characteristics to those observed in Chinese hamster ovary cells expressing CLIC1. J Biol Chem.277: 26003-11.
- Averaimo S,Milton RH,Duchen MR,Mazzanti M. (2010). Chloride intracellular channel 1 (CLIC1): Sensor and effector during oxidative stress. FEBS Lett. 584: 2076-84.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences