Precise Relationship Between Chlamys farreri, Mizuhopecten yessoensis Inferred from Internal Transcribed Spacers and 5.8s Ribosomal DNA
Haidong Tan, Qiu Chen, Linmei Wang, Weidong Liu, Hao Su
1Dalian Institute of Chemical Physics,CAS,116023 Dalian,P.R.China
2Liaoning Sea Fisheries Research Institute,116023 Dalian,P.R.China
3Dalian institute of Biotechnology,Liaoning Agricutural Academy,116023 Dalian,P.R.China
Abstract
ITS2 (Internal Transcribed Spacer2) region and 5.8S, partial 28S of ribosomal DNA was PCR amplified and sequenced in Chlamys farreri, Mizuhopecten yessoensis. For each species, two clones were examined. Alignment of sequence suggested the presence of ITS2 with 272 –286 bp respectively, 5.8S with 155 bp. Both entire 5.8S and partial 28S with 118 bp in length were 100% homologous and ITS2 were responsible for sequence variation between the two pieces while ITS2 was 100% homologous within intraindividual Chlamys farreri and 96.5% homologous within intraindividual Mizuhopecten yessoensis. Presence of sequence variation in the ITS region suggested that it could be used for assaying genetic diversity in scallops. These results provide new insights into precise identification of relationships between scallop species.
Keywords
Chlamys farreri; Mizuhopecten yessoensis; internal transcribed spacers; 5.8s ribosomal DNA
1. Introduction
With the declining of catches on the coast and the increasing in international food trade,there is a higher risk that of incorrect labeling for scallops specimens,so the authentication of scallops has been extremely demanded. In this field accurate species identification is the most daunting task,which requires the presence of the shell and a detailed analysis when specimens are small in size [1] although identification of many scallops is mostly achieved by employing morphologic criteria [2]. For these reasons,molecular biotechnology seems to have many advantages for its effectiveness and simplicity.
Eukaryotic ribosomal RNA genes are found as parts of repeat units that are arranged in tandem arrays. Each repeat unit consists of a transcribed region (having genes for 18S,5.8S and 28S rRNAs and the external transcribed spacers ETS1 and ETS2) and a non-transcribed spacer (NTS) region. In the transcribed region,internal transcribed spacers (ITS) are found on either side of 5.8S rRNA gene and are described as ITS1 and ITS2. The length and sequences of ITS regions of rRNA repeats are believed to be fast evolving and therefore may vary. Universal PCR primers designed from highly conserved regions flanking the ITS and its relatively small size (270-280 bp) enable easy amplification of ITS region due to high copy of rRNA repeats. This makes the ITS region an interesting subject for genetic markers,evolutionary and phylogenetic investigations [3,4] as well as biogeographic investigations [5-7]. In bivalves the 18S rRNA gene has provided genetic markers to distinguish the scallops Placopecten magellanicus and Chlamys islandica [8].
2. Materials and Methods
Chlamys farreri,Mizuhopecten yessoensis were supplied by professor Qiu Chen from Liaoning Sea Fisheries Research Institute,China. Samples were stored in an aqueous solution of 20% (v/v) DMSO saturated with NaCl [9] or kept frozen until DNA extraction. Total genomic DNA was isolated from 50 mg scallop adductor muscle using the CTAB-based protocol [10]. The 5.8S,ITS2 and partial 28S was amplified by PCR using the bilateral conserved regions as the primers (forward,tcactctaagcggtggatcac;gctcttcccgcttcactcgcc). PCR was performed in a 50 ul volume composed of 20 ng genomic DNA,0.2 mM of each dNTP,0.4 uM of each primer,reaction buffer (10 mM Tris.HCl,1.5 mM MgCl2,and 50 mM KCl),and 2 U Taq polymerase. The thermal cycler protocol consisted of an initial denaturation of 2 min at 94°C,followed by 25 cycles of 94°C for 30 s,55°C for 30 s,and 72°C for 1 min,and a final extension for 10 min at 72°C. All amplified fragments were checked by running a sample on 1.5% w/v agarose gel. For one individual from each species,the product obtained from two separate PCR was cloned using the pGEM-T Easy Vector System II (Promega,Madison,WI). After the transformation of Escherichia coli DH5a competent cells,recombinant colonies were selected. Plasmid DNA was purified using miniplasmid preparation kit (Dingguo Biotechnology Ltd,Beijing).The PCR products from 4 scallops (two scallops were selected from each species) were sequenced in Takara Biotechnology. The corresponding nucleotide sequences have been deposited in Genbank under accession numbers AY776156,AY776157,AY860597 (The two former samples had the same sequences in the same accession number).
The identity of the sequences obtained was corroborated using the BLAST program and the boundaries of the coding and spacer regions were determined by comparison with sequence data from the same organisms in Genbank. The alignment was achieved with vector NTI suite 9 (Invitrogen Corporation,Carlsbad,CA).
3. Results
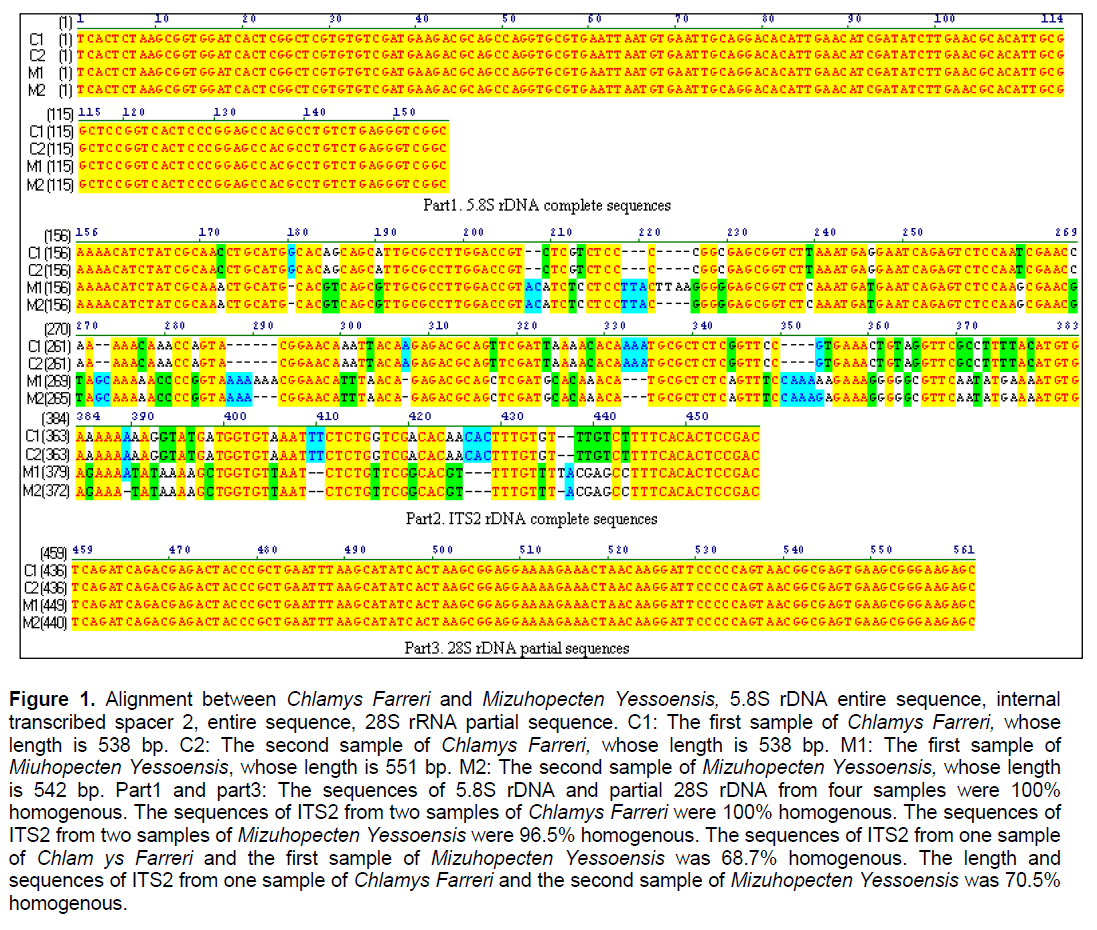
The total length of the entire PCR products was about 538-551 bp. There was no single variable site in 5.8S rDNA and partial 28S rDNA (Figure 1. part 1 and part 3). The results suggested that transcribed regions are highly conserved,even in some different species showed 100% homogeneity,which also implied that Chlamys Farreri and Mizuhopecten yessoensis originated from the closely related progenitor. Other evidences also support the result,such as both of them have a diploid number of 38 chromosomes. However,the variation in the total length and sequences of the entire ITS2 between Chlamys Farreri and Mizuhopecten yessoensis was obvious,even showed intraspecific sequence divergence between two different individuals (Figure 1,part 2,96.5% homologous within intraindividual Mizuhopecten yessoensis). Except the variation,some conserved regions were still existed,such as about 16bp-base region at the left flanking gene of ITS2 and 14bp-base region at the right flanking gene of ITS2 (Figure 1,part 2).
Figure 1. Alignment between Chlamys Farreri and Mizuhopecten Yessoensis, 5.8S rDNA entire sequence, internal transcribed spacer 2, entire sequence, 28S rRNA partial sequence. C1: The first sample of Chlamys Farreri, whose length is 538 bp. C2: The second sample of Chlamys Farreri, whose length is 538 bp. M1: The first sample of Miuhopecten Yessoensis, whose length is 551 bp. M2: The second sample of Mizuhopecten Yessoensis, whose length is 542 bp. Part1 and part3: The sequences of 5.8S rDNA and partial 28S rDNA from four samples were 100% homogenous. The sequences of ITS2 from two samples of Chlamys Farreri were 100% homogenous. The sequences of ITS2 from two samples of Mizuhopecten Yessoensis were 96.5% homogenous. The sequences of ITS2 from one sample of Chlam ys Farreri and the first sample of Mizuhopecten Yessoensis was 68.7% homogenous. The length and sequences of ITS2 from one sample of Chlamys Farreri and the second sample of Mizuhopecten Yessoensis was 70.5% homogenous.
4. Discussion
Chlamys Farreri was native specie and cultured in Liaodong Peninsula,whose rDNA regions were relative stable. Mizuhopecten Yessoensis was purchased from Japan,may be adapted to specific climatic conditions and hence may be genetically more diverse. Speculating on above reasoning,ITS regions should be stable from the same species in local areas,whereas some sites in ITS maybe changed or deleted or inserted when the living parameters changed. Using ITS to distinguish different species could be reasonable for the native species,but should be carefully considered when culturing parameters has changed. ITS detecting would be the most precise methods if the two individuals were high homogeneous. These results provide new insights into precise identification and the evolutionary relationships of scallop species.
5. Acknowledgements
The authors wish to thank Professor Zongbao Zhao for critical reviewing of the manuscript and providing some precious suggestion. This study was supported by Liaoning Sea Fisheries Research Institute.
References
- Pena J.B.,Rios C.,Pena S. and Canales J. (1998) Ultrastructural morphogenesis of pectinid spat from the western Mediterranean: a way to differentiate seven genera. J. Shellfish Res. 17: 123-130.
- Wagner H.P. (1991) Review of the European pectinidae. Vita Marina 41:1-48
- Baldwin B.G.,Sanderson M.J.,Porter J.M.,Wojciechowski M.F.,Campbell C.S. and Donoghue M.J. (1995) The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann. Mo. Bot. Gard. 82: 247-277.
- Hershkovitz M.A.,Zimmer E.A. and Hahn W.J. (1999) Ribosomal DNA sequences and angiosperm systematics. In: P.M. Hollingsworth,R.M. Bateman and R.J. Gornall eds. Molecular systematics and plant evolution. Taylor & Francis,London pp.268-326.
- Suh Y.,Thien L.B.,Reeve H.E. and Zimmer E.A. (1993) Molecular evolution and phylogenetic implications of riboso- mal DNA in Winteraceae. Am. J. Bot. 80: 1042-1055.
- Hsiao C.,Chatterton N.J.,Asay K.H. and Jensen K.B. (1994) Phylogenetic relationships of 10 grass species: an assessment of phylogenetic utility of the internal transcribed spacer region in nuclear ribosomal DNA in monocots. Genome 37: 112-120.
- Dubouzet J.G. and Shinoda K. (1999) Relationships among old and New world Alliums according to ITS DNA sequence analysis. Theor. Appl. Genet. 98: 422-433.
- Kenchington E.,Naidu K.S.,Roddick D.L.,Cook D.L.,and Zouros E. (1993) Use of biochemical genetics markers to discriminate between adductor muscles of the sea scallop (Placopecten magellanicus) and the Iceland scallop (Chlamys islandica). Can. J. Fish Aquat Sci. 50: 1222-1228.
- Amos B.,Hoelzel A.R. (1991) Long-term preservation of whale skin for DNA analysis. In: The genetic ecology of whales and dolphins (ed. Hoelzel AR),Special publication No. 13 of the IWC. Cambridge,England pp. 99-103.
- Winnepenninckx B.,Backeljau T.,De Wachter R. (1993) Extraction of high molecular weight DNA from mollusks. Trends in Genetics 9: 407.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences