PAX5/BSAP Transactivates RAG-mediated Immunoglobulin Gene Recombination
Ti He, Zhixin Zhang
Division of Developmental and Clinical Immunology,Departments of Medicine and Microbiology,University of Alabama at Birmingham,Birmingham,AL 35294,USA
Abstract
Generation of diversified immunoglobulin (Ig) and T cell receptor (TCR) repertoires through V(D)J recombination is the foundation of adaptive immunity. For immunoglobulin heavy chain (IgH) gene, the step of VH to DJH rearrangement occurs exclusively in B lineage cells. Accumulating studies indicate that Pax5 (BSAP), a B lineage specific transcription factor, plays an important role in the regulation of B lineage specific VH to DJH recombination. In Pax5-/- mice, the recombination of upstream VH558 genes is severely impeded. Conversely, ectopic expression of Pax5 transgenes in early thymocytes initiates VH7183 to DJH recombination. Pax5 is essential for IgH locus contraction and histone hypo-methylation across the IgH locus prior to V(D)J recombination. We found that Pax5 associates with the VH gene coding regions and interacts with the RAG1 and RAG2 protein complexes to enhance RAG-mediated VH to DJH recombination. Based on our results, we proposed a novel experimental model that Pax5 directly transactivates RAG-mediated VH to DJH recombination.
Keywords
B cell,Pax5/BSAP,immunoglobulin gene,RAG,V(D)J recombination.
1. Generation of a diversified immunoglobulin gene repertoire through RAG-mediated V(D)J recombination
The function of the adaptive immune system is dependent on the tremendously diversified repertoires of immunoglobulin (Ig) and T cell antigen receptor (TCR). The coding exons of the variable domains of Ig and TCR genes are generated through somatic rearrangement of previously separated variable (V),diversity (D),and joining (J) gene segments in developing B and T lineage cells,respectively [1-6]. The recombination process is catalyzed by a pair of recombination activating gene products,RAG1 and RAG2 [7-9]. Both proteins are required for rearrangement of Ig and TCR genes,and disruption of either gene in mice results in a complete block of B and T cell generation [10,11]. For Ig and TCR genes,specific recombination is directed by the recombination signal sequence (RSS) flanking each V,D,or J gene segment [12]. The RSS is composed of two highly conserved motifs,a heptamer (5’-CACAGTG-3’) and a nonamer (5’-ACAAAAACC-3’),separated by a non-conserved spacer with either 12-bp or 23-bp in length [12]. The heptamer and nonamer are important for RAG complexes binding and cleavage of the RSS [13,14],while the length of the spacer region specifies the recombination of different gene fragments [12]. Known as the 12/23 rule,recombination preferentially occurs between a pair of 12-bp and 23-bp RSS tagged gene segments [12].
2. V(D)J recombination is tightly regulated at multiple steps
To ensure the successful assembly of Ig or TCR genes and mean time,to prevent unwanted cleavage of genomic DNA,V(D)J recombination is tightly controlled at different levels. First,recombinations of Ig and TCR genes are restricted in early developing B and T lineage cells,respectively [5,6]. This developmental stage specific regulation is mainly achieved through regulation of the RAG1 and RAG2 gene expression and the differential accessibilities to the rearranging gene loci. RAG1 and RAG2 gene expression is initiated in the common lymphoid progenitor cells and continued in early B and T lineage cells [15]. In B lineage cells,RAG proteins are expressed in pro B cell stage to catalyze IgH gene DH to JH recombination and then VH to DJH recombination. After generation of functional μ heavy chains to form the pre-B cell receptors (pre-BCR),RAG1 and RAG2 gene expression will be temporarily down modulated in large pre B cells [16,17],to prevent additional DNA cleavage during cell proliferation. After expansion of the pre B cells,RAG genes are reexpressed in the small pre B cells to catalyze immunoglobulin light chain (IgL) gene recombination [16,17]. Functionally rearranged IgL genes will produce conventional light chains to form the B cell receptors (BCR). RAG gene expression can be reinduced in immature B cells to edit IgH and IgL genes encoding self reactive BCRs [18,19] and then,will be shut down in mature B cells.
To prevent unwanted DNA damage during cell proliferation,the V(D)J recombinase activity is strictly confined at the G1 phase of cell cycle through regulation of the RAG2 protein level [20-22]. At the G2/M phases,cell cycle-dependent phosphorylation at Thr490 of RAG2 targets RAG2 to the proteosome-dependent degradation process [20-22]. Overexpression of p27kip blocks cell cycle progression and results in accumulation of RAG2 protein. Mutation of the Thr490 to Ala on RAG2 disrupts its cell cycle dependent degradation and leads to increased aberrant signal joint formation in transgenic mice [23,24].
3. The locus accessibility theory for regulation of V(D)J recombination
To ensure the expression of correct antigen receptors on appropriate cells,V(D)J recombination is tightly controlled in a lineage specific manner [2,5,6]. In developing B and T lymphocytes,there are total seven antigen receptor gene loci,including IgH,Igκ,Igλ,TCRα,TCRβ,TCRδ,and TCRγ,that are subjected to RAG-mediated recombination [2,5,6]. Ig genes are only rearranged in B lineage progenitor cells to generate Ig (BCR); while TCR genes are only assembled in early T lineage cells to produce TCR [2,5,6]. Such lineage specific regulation of V(D)J recombination has been recognized from the very beginning,however,the underlying molecular mechanisms are still unclear. The current understanding of the developmental stage- and lineage- specific regulation of V(D)J recombination is based on the accessibility theory [25-27],which was originally proposed to explain the observation that Ig gene germline transcription precedes V(D)J recombination in progenitor B cells [25,28]. Germline transcript was thus considered as the first indicator for the accessibilities to Ig or TCR loci. The accessibilities to the Igκ or TCRα loci were directly examined in the in vitro cleavage assays using purified RAG proteins and intact nuclei substrates prepared from B cells,T cells,or fibroblasts [29]. RAG-mediated cleavage at the JH RSS only occurs in B lineage nuclei,but not in T lineage and fibroblast nuclei. Conversely,RAG-mediated cleavage at the TCRδ RSS only occurs in T lineage nuclei,but not in other lineage nuclei [29]. These results provided the first evidence that the accessibilities to IgH and TCRδ loci are differentially regulated in B and T lineage cells.
In mammalian cells,DNA is packed into chromatin structure,which prevents most cellular reactions using DNA as templates or substrates,including transcription,V(D)J recombination,and DNA repair [30]. Modification of chromatin structure thus becomes an important mechanism to regulate these reactions. It has long been recognized that the core histone tails can be reversibly acetylated by histone acetyltransferases,such as CBP,p300,PCAF,and GCN5 at multiple lysine residues [31-34]. The chromatin structure can also be remodeled by chromatin remodeling complexes,such as the SWI/SNF complexes [35,36]. The mammalian SWI/SNF complex is a giant 2 mega Dalton complex that contains either the Brg1 or Brm ATPase,with a variable composition of subunits,known as the Brg1-associated factors (BAFs) [35,36]. It has been well documented that histone acetylation and chromatin remodeling are important activators in transcriptional regulation.
The requirement for histone acetylation and chromatin remodeling by SWI/SNF complex during RAG-mediated V(D)J recombination has also been well recognized. In B and T lineage cells,the histone acetylation patterns within the IgH and TCRα loci correlate with their recombination status,respectively [37,38],and thus serve as indicators for locus accessibility. In the in vitro cleavage assays,naked DNA substrates with either 12-bp or 23-bp RSS can be efficiently cleaved by purified RAG1/RAG2 proteins regardless of their Ig or TCR origins [39,40]. However,when the same RSS substrates were reconstituted into nucleosomal structures with purified core histone components,RAG-mediated cleavage was completely blocked [41-43]. Artificial modification of the histone tails by partial trypsin digestion,hyperacetylation,and supplementation with the chromatin remodeling SWI/SNF complexes enhance RAG-mediated cleavage of the chromatinized RSS templates [41,43,44]. The ATPase component of the SWI/SNF chromatin remodeling complex,Brg1,has been found to associate with the antigen receptor gene loci poised for V(D)J recombination,implying that SWI/SNF is involved in remodeling the chromatin structure for V(D)J recombination in vivo [45]. Clearly,histone acetyltransferase and chromatin remodeling complexes are important regulators in RAG-mediated recombination. However,how these generalized chromatin modification factors are specifically recruited to different rearranging genes is not clear.
4. Regulation of Ig gene recombination
Ig gene rearrangement occurs in a step-wised fashion during early stages of B lymphopoiesis. Normally,DH to JH rearrangement occurs first on one of the two IgH alleles,followed by VH to DJH recombination and then light chain gene rearrangements [46-48]. DH to JH rearrangement marks the onset of B lineage differentiation,but this step of recombination is not B lineage specific. DH-JH joints can be detected in thymocytes [49]. The step of VH to DJH rearrangement is B lineage specific. Functionally rearranged IgH genes will produce μ heavy chains (μHC) to form the pre B cell receptors (pre BCR) that define the pre B cell phenotype. Understanding how the B lineage specific VH to DJH recombination is regulated remains an important topic in immunology.
B lineage specific transcription of the IgH gene is controlled by three regulatory elements,the VH gene promoter,the intronic μ enhancer (Eμ),and the 3’ IgH enhancer [50]. A combination of the VH promoter and the Eμ enhancer can successfully direct B lineage specific expression of foreign transgenes. The important role of the Eμ enhancer in the regulation of B lineage specific VH to DJH recombination has been studied extensively in different experimental systems [51-53]. First,deletion of the Eμ enhancer on one of the IgH alleles significantly affects the VH to DJH recombination,but with only mild effect on DH to JH recombination [51,52]. Detailed mapping of the Eμ enhancer showed that the core region without the flanking matrix attachment regions is sufficient to promote normal V(D)J recombination [53]. Second,when placed downstream of a mini TCR locus,the Eμ enhancer can direct specific recombination of this artificial construct in B lineage cells in a transgenic mouse [54]. The function of the VH gene promoter in the regulation of VH to DJH recombination has not been fully studied. Early reports using a chicken Igλ mini locus showed that the promoter was important for the efficient recombination of that construct [55]. Recent studies using the mini TCRα locus showed that the important function of the TCRα promoter is to enhance the accessibility to the recombination constructs rather than to initiate the transcription of the Vα gene,as the promoter can be placed in either orientations relative to the Vα gene [54].
E2A,EBF,and Pax5 are important transcriptional regulators for early B lineage cell development and also play important roles in regulation of Ig gene recombination [56,57]. E2A was originally identified as an Ig enhancer binding factor [58]. The E2A gene encodes two proteins,E12 and E47,through differential RNA splicing. E12 and E47 are involved in the regulation of many B lineage specific genes expression,including RAG1,RAG2,λ5,VpreB,Pax5,and EBF [59,60]. In mice deficient of E2A activity either by targeted disruption of the E2A gene or by forced expression of the E2A inhibitor Id1 protein,B lineage cell development was completely blocked before the stage of DH to JH recombination [61-63]. EBF is a transcriptional regulator that is expressed in various tissues,including olfactory neurons,adipocytes,and B lineage cells [64]. EBF regulates the expression of many B lineage specific genes,including mb-1,B29,λ5,VpreB,RAG1,and Pax5 [57]. In EBF deficient mice,B cell development is arrested at the pro B cell stage before DH to JH recombination [65]. These results indicated that E2A and EBF are dispensable for the initiation of IgH recombination. The activation function of E2A and EBF in IgH recombination is likely through regulating the locus accessibility. Indeed,previous studies have shown that forced expression of E47 activates IgH germline transcription and TdT gene expression in fibroblasts [66] and forced expression of E2A in a pre T cell line initiates IgH D to JH rearrangement [67]. Recent studies have shown that forced expression of E2A or EBF together with the RAG1 and RAG2 genes in human embryonic kidney BOSC cells initiates endogenous IgH DH to JH,Vκ to Jκ,and TCRγ Vγ to Jγ gene rearrangements [68]. These results further confirmed the important function of E2A and EBF in V(D)J recombination. The failures to complete VH to DHJH recombination and to maintain the lineage specific pattern of rearrangement suggest that additional factors are required for the precise regulation of V(D)J recombination.
5. Pax5 (BSAP) is a B lineage specific transcription factor
Pax5 is an important regulator for B lineage cell development and function [56,57]. Pax5 belongs to the PAX family of transcriptional regulators,which contain the conserved paired DNA binding domain [69]. Pax5 also has a partial homeodomain and transcriptional activation and inhibitory domains [70-72]. Pax5 is mainly expressed in B lineage cells and in developing midbrain [69,73]. In B lineage cells,Pax5 expression is initiated in the common lymphoid progenitor cells and extinguished in terminally differentiated plasma cells [69]. In Pax5-/- mice,B lineage cell development is blocked at pro B cell stage [74,75]. Pax5 controls the transcription of many B lineage specific genes,including Cd19,Blk,Mb-1,and Blnk,through binding to their promoters [76]. A detailed comparison of the gene expression profiles between the wild type and the Pax5-/- pro B cells confirmed many previously identified Pax5 target genes and also revealed several new ones [76]. Forced expression of IgH and Igκ transgenes fail to advance Pax5-/- pro B cell development (69). Blnk is another Pax5 target gene,which encodes an important adaptor protein to transduce signals from the pre-BCR and BCR. Loss of Pax5 results in no Blnk expression and defective pre-BCR signaling (70). Reconstitution of Blnk expression in Pax5-/- B cells restored the pre-BCR-mediated signaling,but was not enough to rescue B cell development [77]. These results revealed the sequential requirement for Pax5 and its target genes at different stages of B cell development. However,these results can not fully explain the severe blockade of B cell development in Pax5-/- mice.
Pax5 also acts as a restriction factor to specify the B lineage cell differentiation pathway through suppression the expressions of lineage or differentiation stage non-appropriate genes [78,79]. Without such restrictions,Pax5-/- pro B cells can differentiate into myeloid,T,and dendritic cells [78,80]; and loss of Pax5 in mature B cells also promotes plasma cell differentiation [79,81].
To exert its multiple biological functions,Pax5 interacts with a diverse array of cellular proteins. Pax5 interacts with Ets-1 through its N-terminal region [82,83]. Through this interaction,Ets-1 recruits Pax5 to the Mb-1 gene promoter region,where they bind to a complex Ets-1/Pax5 binding site to enhance transcription [84]. A recent study showed that Pax5 interacts with the Ada2 adaptor protein,GCN5 histone acetyltransferase,and the SWI/SNF chromatin remodelling complex [85]. Through this functional interaction,co-expression of Ada2 and GCN5 dramatically enhances Pax5-mediated transcriptional activation function.
6. Pax5 is required for efficient IgH VH to DJH recombination
An important function of Pax5 in the regulation of IgH VH to DJH recombination was first indicated by the defective VH to DJH recombination in Pax5-/- pro B cells with normal DH to JH recombination [75]. Detailed analyses of the recombination status of different VH genes in Pax5-/- pro B cells revealed that the recombinations of the DH distal VHJ558 genes are more severely compromised than that of the DH proximal VH7183 genes [86]. Surprisingly,the germline transcriptions of VHJ558 and VH7183 genes and the global histone acetylation pattern across the IgH locus,two of the general indicators of locus accessibility,are not affected in Pax5-/- pro B cells [86]. However,without Pax5,the accessible VH genes can not be efficiently rearranged. When the rearranged VH7183 genes in the Pax5-/- pro B cells were sequenced,it becomes clear that even within the VH7183 family,only a few VH genes (including the VH 81X) that are close to the DH locus can be rearranged at normal frequency [87]. The recombinations of upstream VH7183 genes are also defective in Pax5-/- pro B cells [87]. These results suggested that Pax5 is required for the recombination of most VH genes.
Conversely,forced expression of Pax5 in early T lineage cells induced ectopic VH to DJH recombination [88,89]. Interestingly,in these experiments,Pax5 only induces the rearrangement of some DH proximal VH7183 genes but not the upstream VHJ558 genes [88,89]. These results confirmed the important function of Pax5 in the regulation of the VH to DJH recombination and also suggested that additional regulators are required to fully activate B linage specific IgH gene recombination.
Recent studies using 3-dimensional FISH techniques showed that the IgH locus is contracted in normal pro B cells prior to recombination. However,such contraction was not seen in Pax5-/- pro B cells [88]. Pax5 might facilitate the recombination of distal VH genes by contraction the IgH locus to bring upstream VH genes close to the DJH region [87,88]. The IgH locus contraction occurs at an earlier step before DH to JH recombination,because contraction of the IgH locus can be seen in RAG-/- pro B cells and defective IgH locus contraction has also been observed in E2A-/- pro B cells [88,90].
7. Pax5 acts as a transactivator for RAG-mediated VH to DJH recombination
Our recent studies revealed a novel function of Pax5 as a direct activator for RAG-mediated VH to DJH recombination. As a B lineage specific transcription factor,Pax5 exerts most of its biological functions through binding to the cognate DNA binding sites within the promoters or regulatory regions of its target genes [56]. To explore the potential mechanism for how Pax5 regulates VH to DJH recombination,we searched for potential Pax5 binding sites within the VH gene promoters,VH gene coding regions,and 3’ flanking regions of human and mouse VH genes using computer based transcription factor binding site search programs (TESS). Interestingly,our initial sequence analysis identified clustered Pax5 binding sites within the VH gene coding regions of most human and mouse VH genes,but not in the VH gene promoter or the 3’ flanking regions [91]. The Pax5 binding capacities of these VH gene derived Pax5 binding sites were first confirmed by electrophoresis mobility shift assays (EMSA). Pax5 protein in the crude nuclear extracts prepared from B lineage cells,purified his-tagged Pax5 proteins,and purified PRD peptides containing the Pax5 DNA binding domain all bind to individual Pax5 binding sites derived from representative VH genes of human and mouse origins [91]. Although the individual Pax5 binding sites from VH genes display a lower Pax5 binding affinity comparing to the Pax5 binding site from the CD19 gene promoter region,the full length VH gene coding regions have a comparable Pax5 binding affinity to that of the CD19 promoter region containing two high affinity Pax5 binding sites [91].
To determine if Pax5 associates with the VH gene coding regions in developing B lineage cells,we performed chromatin immunoprecipitation (ChIP) assays. Human EU12 cells express endogenous Pax5,RAG1,RAG2,and CD19 genes with ongoing V(D)J recombination and were used in our ChIP assays. Enrichment of the VH1 and VH3 coding regions was found in anti-Pax5 antibody precipitated chromatin samples in real time PCR based analyses [91]. These results confirmed that Pax5 associates with the VH gene coding regions in the EU12 cells. Using pro B cells from the μMT mice,enrichment of the V1,V11,VHJ558,and VH7183 genes was also found in anti-Pax5 antibody precipitated chromatin samples [91],indicating that Pax5 associates with the VH coding regions in mouse early B lineage cells.
The potential function of Pax5 in regulating RAG-mediated recombination was first suggested by the co-immunoprecipitation studies. Using anti-RAG1 or anti-RAG2 antibodies,we can immunoprecipitate Pax5 from nuclear extracts prepared from human EU12 cells,which contains endogenous Pax5,RAG1,and RAG2 proteins [91]. Conversely,using anti-Pax5 antibodies,we can immunoprecipitate RAG1 and RAG2 proteins from the same nuclear extracts [91]. The interaction between Pax5 and the RAG1 and RAG2 protein complex was further confirmed by GST-fusion protein pull-down assays. Using co-purified GST-RAG1 and GST-RAG2 core proteins,we can efficiently pull down [35S] Met labelled full length Pax5 proteins generated from the in vitro transcription and translation coupled system (TNT system,Promega) [91]. Individually purified GST-RAG1 or GST-RAG2 protein only interacts weakly with Pax5. Analyses using different Pax5 truncation constructs further showed that the N-terminal of Pax5 is required for interaction with the RAG1 and RAG2 complexes [91]. The interaction between Pax5 and RAG1,RAG2 has been further confirmed in mammalian one-hybridization assays [91].
To directly test the potential function of Pax5 in RAG-mediated recombination,we performed in vitro cleavage assays using the full length VH1-8 gene coding region as substrates. Co-purified RAG1 and RAG2 core proteins cleave the VH1-8 gene substrates and generate coding and signal end products. The addition of purified Pax5 protein into the in vitro cleavage reaction enhances RAG-mediated cleavage at the 23-bp RSS [91].
The activation function of Pax5 in RAG-mediated VH to DJH recombination was further tested in fibroblast based recombination assays using the pJH289-VH1-8 construct containing the VH1-8 gene coding region with three identified Pax5 binding sites. Overexpression of Pax5 enhances RAG-mediated recombination of the pJH289-VH1-8 substrates as determined by PCR amplification of either the coding joints or the signal joints [91]. The Pax5-mediated enhancement of VH to DJH recombination depends on the integrity of the full length Pax5 protein. Deletion of either the N-terminal region or the C-terminal region of Pax5 affects Pax5-mediated activation function [91]. Importantly,the Pax5-mediated activation of VH to DJH recombination is dependent on the Pax5 binding sites within the VH gene coding region. Mutation of the three Pax5 binding sites within the VH1-8 gene almost completely abolished Pax5-mediated activation of recombination in either NIH3T3 fibroblasts or EU12 cells [91].
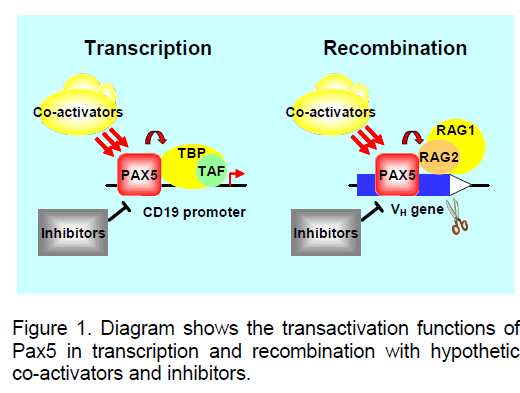
These results led us to propose the current experimental model that Pax5 acts as a transactivator for the B lineage specific IgH VH to DJH recombination (Fig. 1). This novel action of Pax5 parallels closely with its function in transcriptional regulation,in which Pax5 binds to the promoter regions of its target genes and recruits the basal transcriptional machinery and additional co-activators,such as the histone acetyltransferase p300 and the chromatin remodelling SWI/SNF complexes. We speculate that Pax5 binding to the VH gene coding regions could also recruit additional co-activators,such as p300 and the SWI/SNF complexes,to facilitate individual VH gene recombination. This mode of action also predicts that the Pax5 binding capacity to an individual VH gene could directly determine its recombination efficiency and thus influence its usage in the primary IgH repertoire. On the other hand,negative regulators for Pax5 could indirectly regulate VH to DJH recombination and thus provide additional levels of regulation for IgH recombination.
8. Discussion
Collectively,the essential function of Pax5 in regulation of B lineage specific VH to DJH recombination has been well recognized. Currently,two potential mechanisms have been proposed to explain the regulation function of Pax5 in RAG-mediated VH to DJH recombination. Based on the 3-D FISH results,it has been proposed that Pax5 activates IgH gene recombination through contracting the IgH locus. This model provides a suitable explanation for the defective recombination of distal VHJ558 genes in Pax5-/- pro B cells,in that locus contraction could bring the upstream VHJ558 genes closer to the DJH regions and facilitate their recombinations. However,this model can not explain the defective recombination of the upstream VH7183 genes in Pax5-/- pro B cells and the induction of DH proximal VH7183 gene recombination in early T lineage cells expressing Pax5 transgenes.
We proposed the second model that Pax5 acts as a direct transactivator for RAG-mediated VH to DJH recombination. Pax5 binds to the coding regions of individual VH genes and recruits the RAG complexes to enhance recombination,which parallels closely to the activation function of Pax5 in transcriptional regulation. The activation function of Pax5 is required for almost all the VH genes,because most of them have potential Pax5 binding sites. For a few DH proximal VH genes,their recombinations are likely independent of Pax5 in B lineage cells,presumably due to their close proximity to the DJH region and the Eμ enhancer. However,forced expression of Pax5 induces their recombinations in T lineage cells,suggesting that Pax5 also has activation function for these genes.
Thus,despite the different working models,it is reasonable to conclude that the B lineage specific IgH VH to DJH recombination is controlled by the B lineage specific transcription factor,Pax5 (BSAP). These findings will have significant implications for our future studies of the lineage specific regulation of RAG-mediated recombination of other antigen receptor gene loci in developing B and T lineage cells.
Acknowledgments
We thank Dr. Himanshu P. Raikwar for reviewing this manuscript. Z.Z. is supported in part by a K01 grant AR048592 from NIH/NIAMS.
References
- Tonegawa,S. (1983) Somatic generation of antibody diversity. Nature. 302: 575-581.
- Lewis,S.M. (1994) The mechanism of V(D)J joining: lessons from molecular,immunological,and comparative analyses. Adv. Immunol. 56: 27-150.
- Rajewsky,K. (1996) Clonal selection and learning in the antibody system. Nature. 381: 751-758.
- Bassing,C.H.,Swat W.,and Alt F.W. (2002) The mechanism and regulation of chromosomal V(D)J recombination. Cell. 109: S45-S55.
- Jung,D.,and Alt F.W. (2004) Unraveling V(D)J recombination: insights into gene regulation. Cell. 116: 299-311.
- Jung,D.,Giallourakis C.,Mostoslavsky R.,et al. (2006) Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 24: 541-570.
- Oettinger,M.A.,Schatz D.G.,Gorka C.,et al. (1990) RAG-1 and RAG-2,adjacent genes that synergistically activate V(D)J recombination. Science. 248: 1517-1523.
- Schatz,D.G.,and Baltimore D. (1988) Stable expression of immunoglobulin gene V(D)J recombinase activity by gene transfer into 3T3 fibroblasts. Cell. 53: 107-115.
- Schatz,D.G.,Oettinger M.A.,and Baltimore D. (1989) The V(D)J recombination activating gene,RAG-1. Cell. 59: 1035-1048.
- ]Mombaerts,P.,Iacomini J.,Johnson R.S.,et al. (1992) RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 68: 869-877.
- Shinkai,Y.,Rathbun G.,Lam K.P.,et al. (1992) RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 68: 855-867.
- Lewis,S.M. (1994) The mechanism of V(D)J joining: lessons from molecular,immunological,and comparative analyses. Adv. Immunol. 56: 27-150.
- Swanson,P.C.,and Desiderio S. (1998) V(D)J recombination signal recognition: distinct,overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 9:115-125.
- Swanson,P.C.,and Desiderio S. (1999) RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol Cell Biol. 19: 3674-3683.
- Igarashi,H.,Gregory S.C.,Yokota T.,et al. (2002) Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 17:117-130.
- Wang,Y.H.,Stephan R.P.,Scheffold A.,et al. (2002) Differential surrogate light chain expression governs B-cell differentiation. Blood. 99: 2459-2467.
- Burrows,P.D.,Stephan R.P.,Wang Y.H.,et al. (2002) The transient expression of pre-B cell receptors governs B cell development. Semin. Immunol. 14: 343-349.
- Melamed,D.,and Nemazee D. (1997) Self-antigen does not accelerate immature B cell apoptosis,but stimulates receptor editing as a consequence of developmental arrest. Proc. Natl Acad Sci. U. S. A. 94: 9267-9272.
- Sandel,P.C.,and Monroe J.G. (1999) Negative selection of immature B cells by receptor editing or deletion is determined by site of antigen encounter. Immunity. 10: 289-299.
- Lin,W.C.,and Desiderio S. (1993) Regulation of V(D)J recombination activator protein RAG-2 by phosphorylation. Science. 260: 953-959.
- Lin,W.C.,and Desiderio S. (1994) Cell cycle regulation of V(D)J recombination-activating protein RAG-2. Proc. Natl Acad Sci. U. S. A. 91: 2733-2737.
- Lin,W.C.,and Desiderio S. (1995) V(D)J recombination and the cell cycle. Immunol. Today. 16: 279-289.
- Li,Z.,Dordai D.I.,Lee J.,et al. (1996) A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity. 5: 575-589.
- Ross,A.E.,Vuica M.,and Desiderio S. (2003) Overlapping signals for protein degradation and nuclear localization define a role for intrinsic RAG-2 nuclear uptake in dividing cells. Mol Cell Biol. 23: 5308-5319.
- Yancopoulos,G.D.,and Alt F.W. (1985) Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 40: 271-281.
- Sleckman,B.P.,Gorman J.R.,and Alt F.W. (1996) Accessibility control of antigen-receptor variable-region gene assembly: role of cis-acting elements. Annu. Rev. Immunol. 14: 459-481.
- Sleckman,B.P.,Bassing C.H.,Bardon C.G.,et al. (1998) Accessibility control of variable region gene assembly during T-cell development. Immunol. Rev. 165: 121-130.
- Schlissel,M.S.,and Baltimore D. (1989) Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 58: 1001-1007.
- Stanhope-Baker,P.,Hudson K.M.,Shaffer A.L.,et al. (1996) Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 85:887-897.
- Roth,D.B.,and Roth S.Y. (2000) Unequal access: regulating V(D)J recombination through chromatin remodeling. Cell. 103: 699-702.
- Ogryzko,V.V.,Schiltz R.L.,Russanova V.,et al. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 87: 953-959.
- Bannister,A.J.,and Kouzarides T. (1996) The CBP co-activator is a histone acetyltransferase. Nature. 384: 641-643.
- Blanco,J.C.,Minucci S.,Lu J.,et al. (1998) The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 12: 1638-1651.
- Eberharter,A.,and Becker P.B. (2002) Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 3: 224-229.
- Fry,C.J.,and Peterson C.L. (2001) Chromatin remodeling enzymes: who's on first? Curr. Biol. 11: R185-R197.
- Fry,C.J.,and Peterson C.L. (2002) Transcription. Unlocking the gates to gene expression. Science. 295: 1847-1848.
- McMurry,M.T.,and Krangel M.S. (2000) A role for histone acetylation in the developmental regulation of V(D)J recombination. Science. 287: 495-498.
- Johnson,K.,Angelin-Duclos C.,Park S.,et al. (2003) Changes in histone acetylation are associated with differences in accessibility of V(H) gene segments to V-DJ recombination during B-cell ontogeny and development. Mol. Cell Biol. 23: 2438-2450.
- Eastman,Q.M.,Leu T.M.,and Schatz D.G. (1996) Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 380: 85-88.
- Ramsden,D.A.,Paull T.T.,and Gellert M. (1997) Cell-free V(D)J recombination. Nature. 388: 488-491.
- Kwon,J.,Imbalzano A.N.,Matthews A.,et al. (1998) Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Mol. Cell. 2: 829-839.
- Golding,A.,Chandler S.,Ballestar E.,et al. (1999) Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. EMBO J. 18: 3712-3723.
- Kwon,J.,Morshead K.B.,Guyon J.R.,et al. (2000) Histone acetylation and hSWI/SNF remodeling act in concert to stimulate V(D)J cleavage of nucleosomal DNA. Mol. Cell. 6: 1037-1048.
- Patenge,N.,Elkin S.K.,and Oettinger M.A. (2004) ATP-dependent remodeling by SWI/SNF and ISWI proteins stimulates V(D)J cleavage of 5 S arrays. J Biol Chem. 279: 35360-35367.
- Morshead,K.B.,Ciccone D.N.,Taverna S.D.,et al. (2003) Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc. Natl Acad Sci. USA. 100: 11577-11582.
- Burrows,P.,LeJeune M.,and Kearney J.F. (1979) Evidence that murine pre-B cells synthesise µ heavy chains but no light chains. Nature. 280: 838-840.
- Alt,F.,Rosenberg N.,Lewis S.,et al. (1981) Organization and reorganization of immunoglobulin genes in A-MULV-transformed cells: rearrangement of heavy but not light chain genes. Cell. 27: 381-390.
- Alt,F.W.,Yancopoulos G.D.,Blackwell T.K.,et al. (1984) Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 3:1209-1219.
- Kurosawa,Y.,von Boehmer H.,Haas W.,et al. (1981) Identification of D segments of immunoglobulin heavy-chain genes and their rearrangement in T lymphocytes. Nature. 290: 565-570.
- Grosschedl,R.,and Baltimore D. (1985) Cell-type specificity of immunoglobulin gene expression is regulated by at least three DNA sequence elements. Cell. 41: 885-897.
- Chen,J.,Young F.,Bottaro A.,et al. (1993) Mutations of the intronic IgH enhancer and its flanking sequences differentially affect accessibility of the JH locus. EMBO J. 12: 4635-4645.
- Serwe,M.,and Sablitzky F. (1993) V(D)J recombination in B cells is impaired but not blocked by targeted deletion of the immunoglobulin heavy chain intron enhancer. EMBO J. 12: 2321-2327.
- Sakai,E.,Bottaro A.,Davidson L.,et al. (1999) Recombination and transcription of the endogenous Ig heavy chain locus is effected by the Ig heavy chain intronic enhancer core region in the absence of the matrix attachment regions. Proc. Natl Acad Sci. U. S. A. 96: 1526-1531.
- Sikes,M.L.,Meade A.,Tripathi R.,et al. (2002) Regulation of V(D)J recombination: a dominant role for promoter positioning in gene segment accessibility. Proc. Natl Acad Sci. USA. 99: 12309-12314.
- Lauster,R.,Reynaud C.A.,Martensson I.L.,et al. (1993) Promoter,enhancer and silencer elements regulate rearrangement of an immunoglobulin transgene. EMBO J. 12: 4615-4623.
- Schebesta,M.,Heavey B.,and Busslinger M. (2002) Transcriptional control of B-cell development. Curr. Opin. Immunol. 14: 216-223.
- aier,H.,and Hagman J. (2002) Roles of EBF and Pax-5 in B lineage commitment and development. Semin. Immunol. 14: 415-422.
- Murre,C.,McCaw P.S.,and Baltimore D. (1989) A new DNA binding and dimerization motif in immunoglobulin enhancer binding,daughterless,MyoD,and myc proteins. Cell. 56: 777-783.
- Kee,B.L.,Quong M.W.,and Murre C. (2000) E2A proteins: essential regulators at multiple stages of B-cell development. Immunol. Rev. 175: 138-149.
- Greenbaum,S.,and Zhuang Y. (2002) Identification of E2A target genes in B lymphocyte development by using a gene tagging-based chromatin immunoprecipitation system. Proc. Natl Acad Sci. USA. 99: 15030-15035.
- Bain,G.,Maandag E.C.,Izon D.J.,et al. (1994) E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 79: 885-892.
- Sun,X.H. (1994) Constitutive expression of the Id1 gene impairs mouse B cell development. Cell. 79: 893-900.
- Zhuang,Y.,Soriano P.,and Weintraub H. (1994) The helix-loop-helix gene E2A is required for B cell formation. Cell. 79: 875-884.
- Hagman,J.,Belanger C.,Travis A.,et al. (1993) Cloning and functional characterization of early B-cell factor,a regulator of lymphocyte-specific gene expression. Genes Dev. 7: 760-773.
- Lin,H.,and Grosschedl R. (1995) Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 376: 263-267.
- Choi,J.K.,Shen C.P.,Radomska H.S.,et al. (1996) E47 activates the Ig-heavy chain and TdT loci in non-B cells. EMBO J. 15: 5014-5021.
- Schlissel,M.,Voronova A.,and Baltimore D. (1991) Helix-loop-helix transcription factor E47 activates germ-line immunoglobulin heavy-chain gene transcription and rearrangement in a pre-T-cell line. Genes Dev. 5: 1367-1376.
- Romanow,W.J.,Langerak A.W.,Goebel P.,et al. (2000) E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Mol. Cell. 5: 343-353.
- Barberis,A.,Widenhorn K.,Vitelli L.,et al. (1990) A novel B-cell lineage-specific transcription factor present at early but not late stages of differentiation. Genes Dev. 4: 849-859.
- Czerny,T.,Schaffner G.,and Busslinger M. (1993) DNA sequence recognition by Pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev. 7: 2048-2061.
- Czerny,T.,and Busslinger M. (1995) DNA-binding and transactivation properties of Pax-6: three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5). Mol. Cell. Biol. 15: 2858-2871.
- Dorfler,P.,and Busslinger M. (1996) C-terminal activating and inhibitory domains determine the transactivation potential of BSAP (Pax-5),Pax-2 and Pax-820. EMBO J. 15: 1971-1982.
- Adams,B.,Dorfler P.,Aguzzi A.,et al. (1992) Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes,the developing CNS,and adult testis. Genes Dev. 6: 1589-1607.
- Urbanek,P.,Wang Z.Q.,Fetka I.,et al. (1994) Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 79: 901-912.
- Nutt,S.L.,Urbanek P.,Rolink A.,et al. (1997) Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 11: 476-491.
- Nutt,S.L.,Morrison A.M.,Dorfler P.,et al. (1998) Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO J. 17: 2319-2333.
- Schebesta,M.,Pfeffer P.L.,and Busslinger M. (2002) Control of pre-BCR signaling by Pax5-dependent activation of the BLNK gene. Immunity. 17: 473-485.
- Nutt,S.L.,Heavey B.,Rolink A.G.,et al. (1999) Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 401: 556-562.
- Delogu,A.,Schebesta A.,Sun Q.,et al. (2006) Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 24: 269-281.
- Rolink,A.G.,Nutt S.L.,Melchers F.,et al. (1999) Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature. 401: 603-606.
- Nera,K.P.,Kohonen P.,Narvi E.,et al. (2006) Loss of Pax5 promotes plasma cell differentiation. Immunity. 24: 283-293.
- Fitzsimmons,D.,Hodsdon W.,Wheat W.,et al. (1996) Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 10: 2198-2211.
- Fitzsimmons,D.,Lutz R.,Wheat W.,et al. (2001) Highly conserved amino acids in Pax and Ets proteins are required for DNA binding and ternary complex assembly. Nucleic Acids Res. 29: 4154-4165.
- Garvie,C.W.,Hagman J.,and Wolberger C. (2001) Structural studies of Ets-1/Pax5 complex formation on DNA. Mol. Cell. 8: 1267-1276.
- Barlev,N.A.,Emelyanov A.V.,Castagnino P.,et al. (2003) A Novel Human Ada2 Homologue Functions with Gcn5 or Brg1 To Coactivate Transcription. Mol Cell Biol. 23:6944-6957.
- Hesslein,D.G.,Pflugh D.L.,Chowdhury D.,et al. (2003) Pax5 is required for recombination of transcribed,acetylated,5' IgH V gene segments. Genes Dev. 17: 37-42.
- Roldan,E.,Fuxa M.,Chong W.,et al. (2005) Locus 'decontraction' and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat. Immunol. 6: 31-41.
- Fuxa,M.,Skok J.,Souabni A.,et al. (2004) Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 18: 411-422.
- Hsu,L.Y.,Liang H.E.,Johnson K.,et al. (2004) Pax5 activates immunoglobulin heavy chain V to DJ rearrangement in transgenic thymocytes. J Exp Med. 199: 825-830.
- Sayegh,C.,Jhunjhunwala S.,Riblet R.,et al. (2005) Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 19: 322-327.
- Zhang,Z.,Espinoza C.R.,Yu Z.,et al. (2006) Transcription factor Pax5 (BSAP) transactivates the RAG-mediated V(H)-to-DJ(H) rearrangement of immunoglobulin genes. Nat Immunol. 7: 616-624.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences