Natural Compounds Targeting Transforming Growth Factor-ÃÆà ½Ãâò: In Silico and In Vitro Study
Pushpendra Singh, Felix Bast, Ravi Shankar Singh
Pushpendra Singh1,*, Felix Bast1, Ravi Shankar Singh2
1Centre for Biosciences, School of Basic and Applied Sciences, Central University of Punjab, Bathinda, Punjab, India
2Department of Molecular Medicine & Biotechnology, Sanjay Gandhi Post Graduate Institute of Medical Sciences, Lucknow, India.
Received date: September 21, 2016; Accepted date: November 29, 2016; Published date: December 06, 2016
Citation: Singh P, Bast F, Singh RS. Natural Compounds Targeting Transforming Growth Factor-β: In Silico and In Vitro Study. Electronic J Biol, 13:1
Abstract
Inhibition of the tumor-promoting effects of transforming growth factor beta receptor (TGFβR) in carcinogenesis provides a better therapeutic intervention. Various natural compounds, inhibitors of TGFβR have been used for in vitro and in vivo anticancer study. Although very few TGFβR inhibitors are now intensifying in preclinical studies. In this study our aim to investigate TGFβR1, TGFβR2 and TAK1 inhibitor by using molecular docking and in vitro study. Our result revealed that some compounds have better docking energy. Moreover, the effect of two lead molecules epigallocatechin gallate (EGCG) and myricetin on the mRNA expression of TGFβR1 was reported after the 48 hrs treatments in HepG2 and PC3 cancer cell lines. The RT-PCR showed that compound EGCG and myricetin reduced the mRNA expression of TGFβR1 at 80 μM concentration. This molecular docking study provides a better understanding of binding of compounds to the active site of proteins and to summarize the various binding energy, hydrophobic, hydrogen, an electrostatic bond that are decisive for the protein-ligand interactions. Further experimental work will be required for validation of our results.
Keywords
Natural compounds; Cancer; Maestro 9.6; Molecular docking; In vitro.
1. Introduction
The Transforming Growth Factor β (TGFβ) family have a group of multifunctional regulatory proteins that balance a vast variety of physiological functions including cell proliferation, cell cycle arrest and apoptosis. TGFβR1 53kDa protein is also known as activin encoded by TGFβR1 human gene. TGFβ activated protein kinase 1 (TAK1) was acknowledged for protein kinase action that invigorating by TGFβ signaling transduction [1]. Furthermore, it is also demonstrated that TAK1 cytokine play a significant role in inflammation. TGFβ initiates signals by binding to the TGFβR2 and stabilizes the heteromeric complex with the TGFβR1. As a result, TGFβR1 is trans-phosphorylated and activated by TGFβR2 [2,3]. The activated TGFβ1R then propagates the signals through interaction with receptor-associated Smads [4]. Deregulations of TGFβ signaling pathway responsible for cancer initiation and progression and interrupting the tumor promoter properties of TGFβ signaling would be an attractive therapeutic strategy [5-7]. Monoclonal antibodies and small molecule inhibitors that target TGFβ signaling are the most notable strategy have been used in preclinical studies, but they did not prove much promise as anticancer drugs owing to multifaceted roles of TGFβ signaling [8,9].
In recent years, natural compounds have been investigated intensely in the clinical trial of ovarian, breast, cervical, pancreatic and prostate cancers [10]. A number of propitious agents are in clinical development based on discriminatory activity against anti-cancer molecular targets currently being developed. In this context, we studied TGFβ signaling inhibitors (TGFβR1, TGFβR2, and TAK1) from natural sources using in silico and in vitro approaches.
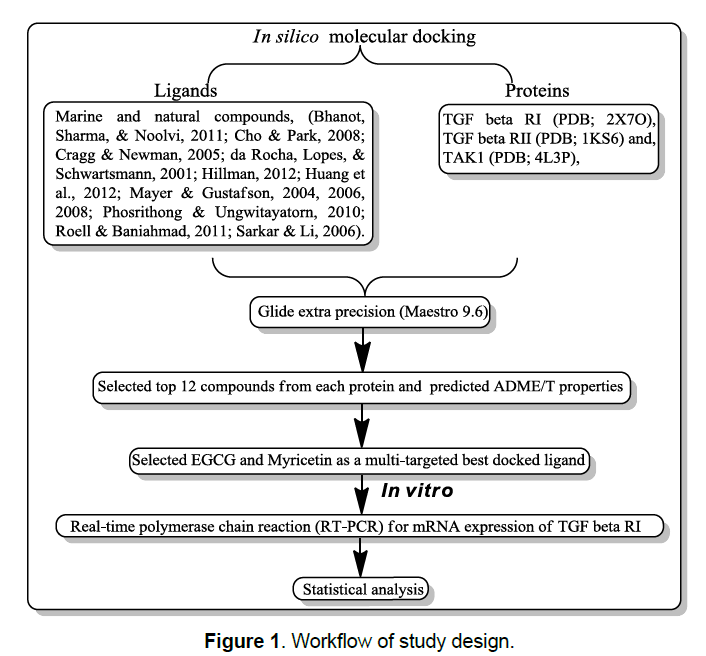
2. Material and Methods
2.1 Selection of ligands and protein molecules
In silico, molecular docking protocol and standard setting of parameter adapted from our published literature [11-16]. Marine and natural compounds selected as ligand molecules that had been reported as an anticancer activity in the published literature [17-29]. The X-ray crystal structure of TGFβR1 (PDB; 2X7O), TGFβR2 (PDB; 1KS6), and TAK1 (PDB; 4L3P) kinase protein retrieved from the Protein Data Bank (PDB) (Figure 1).
2.2 Preparation of ligands molecules
Input ligand molecules were prepared using ligprep wizard applications where the addition of hydrogen atoms, 2D to 3D conversion followed by optimized potential for liquid simulations (OPLS_2005) force field was applied [30-32]. Finally, ten conformations for each ligand were generated and performed molecular docking.
2.3 Preparation of protein molecules
Protein molecules were prepared using respective wizard applications (standard methods) where changes such as the addition of hydrogen atoms, fixing of the charges and orientation of groups, assigning bond orders, creation of disulphide bonds, was done into the PDB structure. After the completion of ligands and protein preparation, a receptor-grid file was generated.
2.4 GLIDE (Grid-based Ligand Docking with Energetics) molecular docking
The active site (binding pocket) and functional residues of protein were identified and characterized by GLIDE from Schrodinger package. GLIDE-XP Molecular docking studies using the selected ligand molecules were conducted using Maestro 9.6 [33- 35]. Each of these selected compounds was docked into target protein molecules and a compound possessing the lowest energy was selected. After the configuration of receptor-grid file, flexible ligands with rigid receptor molecular docking were performed. The concluding energy evaluation was done on the basis of G-score.
2.5 Absorption, distribution, metabolism, excretion and toxicity (ADME/T) studies
ADME properties such as poor compound solubility, gastric emptying time and lack of ability to permeate the intestinal wall can all diminish the extent to which a drug is absorbed after oral administration. Therefore, in silico ADME/T predictive tools are accommodating approach that could abolish unfortunate compounds, before invested valuable time and money in primary testing of compounds. QikProp application of Maestro 9.6 predicts properties such as logBB, overall CNS activity, Caco-2, and MDCK cell permeability and logKhsa for human serum albumin binding, etc. [30- 36].
2.6 Reagents and cell lines culture
DMEM media, 1% penicillin, streptomycin and Fetal Bovine Serum (FBS) procured from Invitrogen, and Ham's F-12 media from HiMedia. EGCG and myricetin purchased from MP Biomedicals Pvt. Ltd. and dissolved in Dimethyl Sulfoxide (DMSO) as a 20 mM stock solution. We used final DMSO concentration in culture medium were 0.25% (vol/vol). Hepatocellular carcinoma (HepG2) and Prostate Cancer (PC3), cell lines were procured from NCCS, Pune, India. Cells were grown in phenol red media containing 10% FBS and 1% penicillin and streptomycin and have to grow in a 37°C incubator with 5% CO2.
2.7 Total RNA isolation, cDNA synthesis and quantitative RT-PCR
One million cells/well were plated into the six-well culture plate media supplemented with 10% FBS and 1% penicillin/streptomycin then incubated at 37°C overnight with 5% CO2. Cells were exposed to 80 μM EGCG and Myricetin for 48 h. After 48 h total RNA was extracted using Tri reagent (Life Technologies, Gaithersburg, MD) according to manufacturer's instruction. RNA was quantified at 260/280 nm absorbance and agarose gel electrophoresis. Total RNA (2 μg) was used to synthesize cDNA using High Capacity cDNA Reverse Transcription Kits (Applied Biosystems) using random hexamers in a final volume of 20 μl as described by the manufacturer. RT-PCRs using SYBR green reagent with genespecific primers (listed in Table 1) and GAPDH gene primers (internal control) were performed in duplicate in a final volume of 20 μl in an ABI 7900 real-time PCR thermal cycler (Applied Biosystems). Relative gene expression levels were calculated after normalization with internal control GAPDH gene using the 2-δδCt method, where Ct is the threshold value. Results were expressed as fold over control.
3. Statistical Analysis
Statistical values are expressed as the mean ± SEM (SigmaPlot). The value of p<0.05 was considered statistically significant.
4. Results and Discussion
4.1 Molecular docking of compounds with TGFβR1, TGFβR2 and TAK1
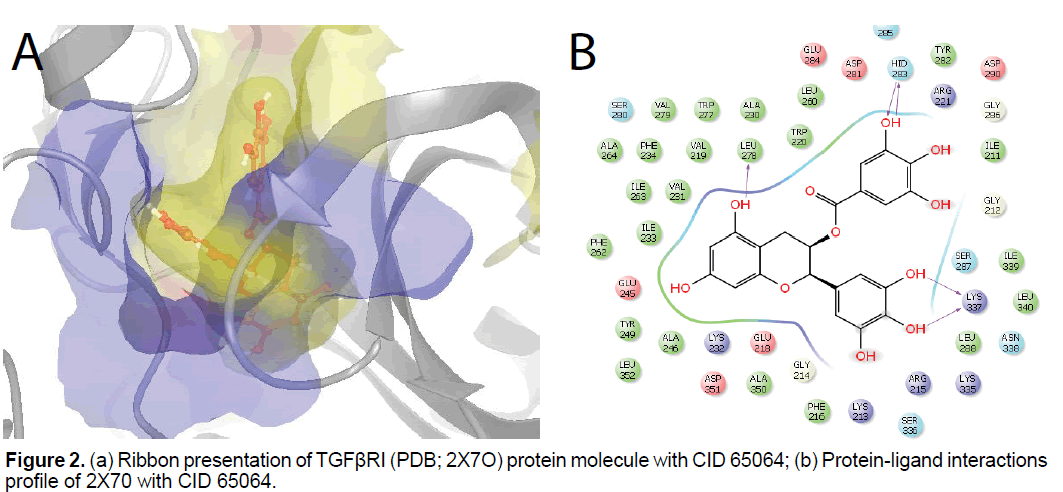
Molecular docking result of TGFβR1 (PDB; 2X7O) against natural compounds revealed compound CID65064 (EGCG), CID72277, CID5281672 (myricetin), CID9892144, CID72276, CID72281, CID5280343, CID9064, CID6445533, CID5280445, CID44259, CID5281614, CID65064 have better Gscore (as compared to control) -13.05, -11.6, -11.36, -11.14, -11.09, -10.94, -10.93, -10.58, -10.47, -10.42, -10.3, -9.98 Kcal/mol, respectively. Moreover, amino acids Ile211, Val219, Val231, Ala230, Tyr249, Leu260, Val279, Phe262, Ile339, Leu340, and Ala360 involved in the hydrophobic interaction at the active site of the protein. Furthermore, it has been shown that amino acids Leu278, Hid183 and Lys337 form back-bone hydrogen bond at the active site of protein with EGCG. Additionally, it has shown that amino acids Lys232 and Glu245 involved in sidechain hydrogen bonding, in addition, to back-bone hydrogen bonding (Hid283) at the active site of TGFβR1 with myricetin (Figure 2 and Table 2).
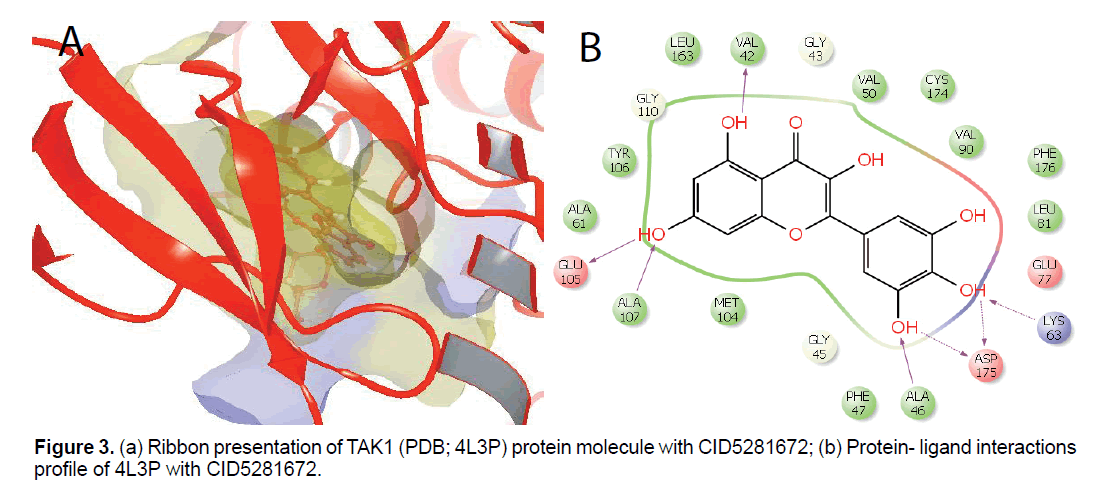
Molecular docking result of TGFβR2 against natural compounds revealed compound CID72276, CID5280343, CID9064, CID5281614, CID65064 (EGCG), CID42607750, CID6400741, CID5281672 (myricetin) have better G-score -8.03, -7.81, -7.71, -7.57, -7.3, -6.98, -6.42, -6.36 Kcal/mol, respectively. Further, it was observed that amino acids Gln41, Lys42, Trp65, Met100 and Phe126 form hydrophobic, side chain hydrogen, backbone hydrogen, π-cation and π-π interactions during protein-ligand interactions of TGFβR2 with EGCG. Consequently, it was shown that amino acids Gln41, Trp65, Met100, Glu128 and Phe126 form hydrophobic, side chain hydrogen, backbone hydrogen, π-cation, and π-π interactions during protein-ligand interactions of TGFβR2 with myricetin. Moreover, molecular docking result of TAK1 against natural compounds revealed compound CID42607750, 5281672, 6445533, 72277, 5280445, 5280343, 9892144, 5281614, 442793, 5281670, 9064 have better G-score (as compared to control) -12.01, -11.67, -11.07, -10.66, -10.2, -10.12, -9.96, -9.42, -9.3, -9.08, -8.87 Kcal/mol, respectively. Moreover, protein-ligand interactions showed that amino acids Phe47, Ala61, Leu81, Val90, Met104, Tyr106, Ala107, Cys174 and Phe176 involved in hydrophobic interaction at the active site of the protein. Moreover, Additionally, it have shown that amino acids Val42, Ala 46, Glu105 and Ala107, involved in side-chain hydrogen bonding whereas amino acid Lys63 and Asp175 involved in back-bone hydrogen bonding at the active site of TAK1 with myricetin. Additionally amino acid Arg44 and Ala107 of TAK1 form backbone hydrogen bonding with EGCG (Figure 3 and Table 3).
| S. No. | Name of gene | Forward primer (5’ to 3’) | Reverse primer (5’ to 3’) |
|---|---|---|---|
| 1. | TGFßRI | 5'-TGCACATCGTCCTGTGGAC-3' | 5'-GTCTCAAACTGCTCTGAAGTGTTC-3' |
| 2. | GAPDH | ACGGATTTGGTCGTATTGGGCG | CTCCTGGAAGATGGTGATGG |
Table 1. Primer sets used for amplification.
| Ligand type | Compounds ID | G-Score | Lipophilic Evdw | HBond | Electro | Protein-ligands interactions |
|---|---|---|---|---|---|---|
| SD 208 | -9.95 | -5.15 | -0.7 | -0.35 | Hid283 | |
| R 268712 | -8.04 | -5.5 | -1.2 | -0.9 | Lys232, Ser287, Asp290 | |
| CID4521392 | -7.87 | -5.16 | -0.81 | -0.25 | Hid283, Lys335 | |
| TGFßR | A 83-01 | -7.33 | -6.89 | -0.05 | -0.58 | Lys337 |
| inhibitors | GW 788388 | -7.29 | -6.3 | -0.35 | -0.42 | Lys232 |
| (Controls) | 449054 | -7.23 | -4.73 | -0.32 | -0.36 | Lys232, Hid283 |
| LY 364947 | -7.07 | -5.09 | -0.17 | -0.32 | Lys232 | |
| D 4476 | -7.05 | -5.66 | -0.88 | -0.29 | Lys232, Glu245, Hid183 | |
| CID11655119 | -7.03 | -7.29 | 0 | -0.34 | ||
| SB 525334 | -6.46 | -4.8 | -0.47 | -0.54 | Lys232, Hid183 | |
| CID65064 | -13.05 | -6.2 | -5.13 | -1.36 | Leu278, Hid183, Lys337 | |
| CID72277 | -11.6 | -4.67 | -4.12 | -1.22 | Lys232, Glu245, Hid183, Asp351 | |
| CID5281672 | -11.36 | -5.02 | -3.96 | -0.99 | Lys232, Glu245, Hid283 | |
| TGFßR1 | CID9892144 | -11.14 | -6.83 | -2.8 | -0.54 | Lys213, Lys232, Hid183, Ser287 |
| CID72276 | -11.09 | -4.72 | -3.46 | -1.12 | Lys232, Glu245, Hid183, Asp351 | |
| CID72281 | -10.94 | -4.97 | -2.94 | -0.98 | Lys232, Glu245, Hid183, Asp351 | |
| CID5280343 | -10.93 | -4.98 | -3.53 | -0.99 | Lys232, Glu245, Hid183 | |
| CID9064 | -10.58 | -4.69 | -2.96 | -1.13 | Lys232, Glu245, Hid183, Asp351 | |
| CID6445533 | -10.47 | -5.29 | -3.06 | -1.18 | Arg215, Leu278, Asp281, Asp351 | |
| CID5280445 | -10.42 | -4.94 | -3.04 | -1.01 | Lys232, Glu245, Hid183 | |
| CID44259 | -10.3 | -5.71 | -1.33 | -0.52 | Asp281, Hid283 | |
| CID5281614 | -9.98 | -4.88 | -2.51 | -0.82 | Asp281, Glu245, Asp351 | |
| CID72276 | -8.03 | -3.69 | -3.51 | -0.77 | Gln41, Trp65, Met100, Phe111, Phe126 | |
| CID5280343 | -7.81 | -3.83 | -3.03 | -0.82 | Gln41, Trp65, Met100, Phe111, Phe126 | |
| CID9064 | -7.71 | -3.71 | -3.03 | -0.8 | Gln41, Trp65, Met100, Phe111, Phe126 | |
| TGFßR2 | CID5281614 | -7.57 | -3.81 | -3.03 | -0.76 | Trp65, Met100, Phe126 |
| CID65064 | -7.3 | -3.29 | -3.3 | -1.15 | Gln41, Lys42, Trp65, Met100, Phe126 | |
| CID42607750 | -6.98 | -2.87 | -3.07 | -1.3 | Met100, Lys101, Phe126 | |
| CID6400741 | -6.42 | -3.32 | -2.23 | -1.42 | Gln41, Trp65, Met100, Phe111, Phe126 | |
| CID5281672 | -6.36 | -2.57 | -2.94 | -0.92 | Gln41, Trp65, Met100, Glu128, Phe126 | |
| CID636888 | -6.27 | -4.12 | -2.29 | -0.6 | Gln41, Glu119, Phe126 | |
| CID72277 | -6.25 | -1.99 | -3.87 | -0.9 | Gln41, Trp65, Phe126 | |
| CID5281605 | -5.87 | -2.2 | -2.4 | -0.74 | Gln41, Trp65, Phe126 | |
| CID439260 | -5.78 | -1.93 | -2.4 | -0.85 | Gln41, Phe126 | |
| CID5280343 | -10.93 | -4.98 | -3.53 | -0.99 | Lys232, Glu245, Hid183 | |
| CID9064 | -10.58 | -4.69 | -2.96 | -1.13 | Lys232, Glu245, Hid183, Asp351 | |
| CID6445533 | -10.47 | -5.29 | -3.06 | -1.18 | Arg215, Leu278, Asp281, Asp351 | |
| CID5280445 | -10.42 | -4.94 | -3.04 | -1.01 | Lys232, Glu245, Hid183 | |
| CID44259 | -10.3 | -5.71 | -1.33 | -0.52 | Asp281, Hid283 | |
| CID5281614 | -9.98 | -4.88 | -2.51 | -0.82 | Asp281, Glu245, Asp351 | |
| CID72276 | -8.03 | -3.69 | -3.51 | -0.77 | Gln41, Trp65, Met100, Phe111, Phe126 | |
| CID5280343 | -7.81 | -3.83 | -3.03 | -0.82 | Gln41, Trp65, Met100, Phe111, Phe126 | |
| CID9064 | -7.71 | -3.71 | -3.03 | -0.8 | Gln41, Trp65, Met100, Phe111, Phe126 | |
| TGFßR2 | CID5281614 | -7.57 | -3.81 | -3.03 | -0.76 | Trp65, Met100, Phe126 |
| CID65064 | -7.3 | -3.29 | -3.3 | -1.15 | Gln41, Lys42, Trp65, Met100, Phe126 | |
| CID42607750 | -6.98 | -2.87 | -3.07 | -1.3 | Met100, Lys101, Phe126 | |
| CID6400741 | -6.42 | -3.32 | -2.23 | -1.42 | Gln41, Trp65, Met100, Phe111, Phe126 | |
| CID5281672 | -6.36 | -2.57 | -2.94 | -0.92 | Gln41, Trp65, Met100, Glu128, Phe126 | |
| CID636888 | -6.27 | -4.12 | -2.29 | -0.6 | Gln41, Glu119, Phe126 | |
| CID72277 | -6.25 | -1.99 | -3.87 | -0.9 | Gln41, Trp65, Phe126 | |
| CID5281605 | -5.87 | -2.2 | -2.4 | -0.74 | Gln41, Trp65, Phe126 | |
| CID439260 | -5.78 | -1.93 | -2.4 | -0.85 | Gln41, Phe126 |
Table 2: Lowest binding energy for the ligand- TGFßR1 (PDB; 2X7O) and TGFßR2 (PDB; 4XJJ) protein interactions.
Previously, it was reported that EGCG can inhibit TGFβ induced epithelial-mesenchymal transitions via down-regulation of phosphorylated Smad2 and Erk1/2 in non-small cell lung cancer cells [37]. Moreover, EGCG significantly subdues the HSP27 induction prompted by TGFβ in a dose-dependent manner. EGCG significantly suppressed the TGFβ induced phosphorylation of stress-activated kinases and c-Jun N-terminal kinases [38]. Furthermore, it was noticed that EGCG interacts with TGFβR2 and inhibits the expression of α-smooth muscle actin via the TGFβ-Smad2/3 pathway in human lung fibroblast cells [39,40]. In vivo study showed that myricitrin was noticeably ameliorated the expression of TGFβ1 and alpha-smooth muscle actin that led to antioxidant, anti-inflammatory and antifibrotic activity [41]. Furthermore, in vivo study in streptozotocin with cadmium-induced nephrotoxic diabetic rats unveil that myricetin up regulates the TGFβ1 and downregulate the peroxisome proliferator-activated receptor alpha proteins expression that led to restraints dyslipidemia and renal mesangial cell proliferation [42].
4.2 ADME/T studies
ADME/T (absorption, distribution, metabolism, elimination and toxicity,) properties of lead molecules were assessed through the Qikprop application of Maestro 9.6. EGCG and myricetin have the best Gscore against TGFβR1, TGFβR2, and TAK1. Furthermore, EGCG and myricetin have their excellent QPlogPo/w, QPlogHerg K+ channels, QPlogBB, QPlogKP, QPlogK HSA values that accomplish the Lipinski’s rule of five. However, these compounds do not have excellent QPP Caco, QPP MDCK values and percentage of human oral absorption. Therefore, EGCG and myricetin structural alteration and optimization are required to predict structures that have better QPP Caco, QPP MDCK and rate of human oral absorption activity (Table 4).
4.3 Effect of EGCG and myricetin on TGFβR1 mRNA expression in HepG2 and PC3 cancer cell lines
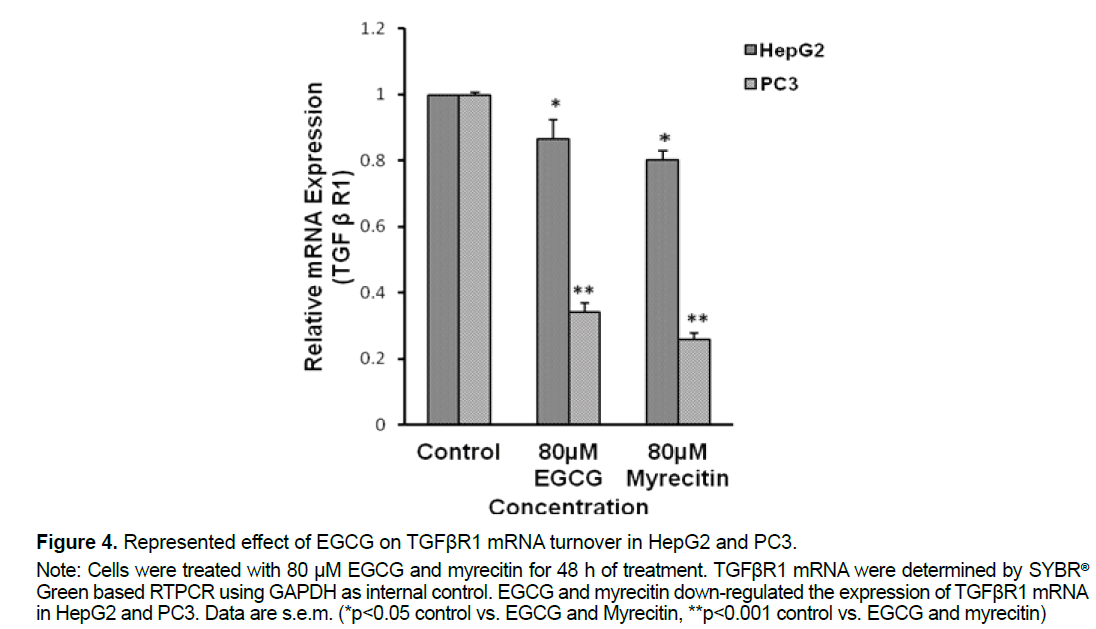
We investigated whether EGCG and myricetin could decrease the TGFβR1 expression in HepG2 and PC3 cells. After 48 h of treatment with EGCG and myricetin, determine the mRNA expression of TGFβR1 by using RT-PCR. As shown in Figure 4, RTPCR demonstrates that EGCG have a prospective for forceful inhibition of TGFβR1. The RT-PCR densitometric bands examination showed that EGCG reduced the mRNA expression of TGFβR1 by 15% in HepG2 (*P<0.05) and 70% (**p<0.001) in PC3 cells at 80 μM concentration. Whereas, RTPCR densitometric bands investigation showed that myricetin reduced the mRNA expression of TGFβR1 by 20% in HepG2 (*P<0.05) and 80% (**p<0.001) in PC3 cells at 80 μM concentrations. TGFβ signaling plays a leading role in the initiation, promotion, and progression of various types of human cancer including prostate, breast lung, and liver cancer. Several lines of evidence suggest that carcinoma cells frequently lose anti-proliferative response to TGFβ. High levels of TGFβ can promote tumor growth in an autocrine and paracrine manner through the stimulation of angiogenesis and metastasis. Blocking the tumor-promoting effects of TGFβ by small molecule inhibitors provides an excellent therapeutic opportunity to improve the treatment of cancer.
5. Conclusion
Figure 4. Represented effect of EGCG on TGFβR1 mRNA turnover in HepG2 and PC3.
Note: Cells were treated with 80 μM EGCG and myrecitin for 48 h of treatment. TGFβR1 mRNA were determined by SYBR®
Green based RTPCR using GAPDH as internal control. EGCG and myrecitin down-regulated the expression of TGFβR1 mRNA in HepG2 and PC3. Data are s.e.m. (*p<0.05 control vs. EGCG and Myrecitin, **p<0.001 control vs. EGCG and myrecitin)
| Ligand type | Compounds CID | G-Score | Lipophilic Evdw | H-Bond | Electro | Protein-ligands interactions |
|---|---|---|---|---|---|---|
| TAK1 Inhibitors | CID9863776 | -6.82 | -3.29 | -2.88 | -0.44 | Ser111, Asn114 |
| CID57369807 | -6.04 | -1.97 | -3.02 | -0.78 | Ser111, Asn114, Asn161 | |
| Natural Compounds | CID42607750 | -12.01 | -5.02 | -4.48 | -1.24 | Arg44, Glu105, Ala107, Ser111, Asn114 |
| CID5281672 | -11.67 | -3.84 | -4.75 | -1.69 | Val42, Ala 46, Lys63, Glu105, Asp175 | |
| CID6445533 | -11.07 | -6.55 | -2.36 | -0.79 | Arg44, Glu105, Ala107, Ser111, Asn114 | |
| CID72277 | -10.66 | -4.62 | -3.83 | -0.82 | Arg44, Glu105, Ala107, Ser111, Asn114 | |
| CID5280445 | -10.2 | -4.25 | -2.95 | -1.57 | Val42, Ala 46, Lys63, Glu105, Asp175 | |
| CID5280343 | -10.12 | -3.81 | -3.5 | -1.53 | Val42, Gly43, Lys63, Glu105, Asp175 | |
| CID9892144 | -9.96 | -6.97 | -2.45 | -0.58 | Ala107, Ser111 | |
| CID5281614 | -9.42 | -4.35 | -3.3 | -1.34 | Val42, Ala 46, Glu105, Ala107, Asp175 | |
| CID442793 | -9.3 | -5.04 | -2.42 | -1.03 | Lys63, Glu105, Ala107, Asp175 | |
| CID5281670 | -9.08 | -5.16 | -2.71 | -0.78 | Lys63, Ala107, Asp175 | |
| CID9064 | -8.87 | -5.11 | -2.46 | -0.89 | Val42, Arg44, Ala107, Asp175 | |
| CID65064 | -8.87 | -4.14 | -4.41 | -0.96 | Arg44, Asp175 |
Table 3: Lowest binding energy for the ligand- TAK1 (PDB; 4L3P) protein interactions.
| Compounds | QPlog Po/w (-2.0 to 6.5) | QPlog HERG (acceptable range: above -5.0) | QPP Caco (nm/s) <25-poor >500- great |
QPlog BB (-3-1.2) |
QPP MDCK (nm/s) <25-poor >500- great |
QPlog Kp (-8.0 to -0.1) |
QPlog Khsa (Acceptable range: -1.5 to 1.5). |
Percentage of human oral absorption; (<25% is poor and >80% is high) |
|---|---|---|---|---|---|---|---|---|
| CID65064 (EGCG) | -0.233 | -5.558 | 1.14 | -4.177 | 0.326 | -7.415 | -0.437 | 0.684 |
| CID72277 | -0.167 | -4.684 | 22.042 | -2.338 | 8.009 | -5.482 | -0.562 | 37.049 |
| CID5281672 (myricetin) | -0.276 | -4.894 | 7.673 | -2.827 | 2.56 | -6.301 | -0.493 | 28.209 |
| CID9892144 | 3.838 | -5.92 | 123.998 | -1.804 | 51.813 | -3.591 | 0.723 | 86.885 |
| CID72276 | 0.479 | -4.852 | 60.119 | -1.858 | 23.692 | -4.599 | -0.43 | 61.589 |
| CID72281 | 1.782 | -5.025 | 133.02 | -1.525 | 55.9 | -4.057 | 0.007 | 75.395 |
| CID5280343 | 0.384 | -5.011 | 20.522 | -2.331 | 7.414 | -5.442 | -0.348 | 52.678 |
| CID9064 | 0.439 | -4.845 | 52.516 | -1.913 | 20.471 | -4.716 | -0.429 | 60.305 |
| CID6445533 | 1.744 | -6.907 | 35.048 | -1.764 | 14.628 | -4.913 | -0.194 | 64.802 |
| CID5280445 | 0.973 | -5.14 | 41.633 | -1.975 | 15.927 | -4.849 | -0.187 | 61.625 |
| CID44259 | 3.765 | -6.13 | 304.782 | -0.05 | 151.525 | -4.131 | 0.787 | 93.45 |
| CID5281614 | 0.505 | -5.178 | 45.418 | -1.943 | 17.497 | -4.758 | -0.364 | 59.565 |
| CID42607750 | -0.607 | -5.487 | 12.878 | -3.215 | 4.48 | -5.266 | -0.883 | 17.34 |
| CID5281614 | 0.458 | -4.924 | 53.31 | -1.808 | 20.806 | -4.65 | -0.388 | 60.532 |
| CID442793 | 3.271 | -4.52 | 506.992 | -1.443 | 237.406 | -2.569 | 0.154 | 94.512 |
| CID9571127 | 2.291 | -4.098 | 0.079 | -4.908 | 0.03 | -6.406 | -0.439 | 0 |
| CID23663412 | 3.425 | -4.65 | 8.604 | -4.303 | 3.837 | -3.649 | -0.49 | 37.809 |
QPlog Po/w (-2.0 to 6.5) Predicted octanol/water partition coefficient; QPlog Herg (acceptable range: above -5.0) Predicted IC50 value for blockage of HERG K+ channels; QPPCaco (nm/sec) <25-poor >500- great Predicted apparent Caco-2 cell permeability in nm/s. Caco-2 cells is a model for the gut blood barrier; QPlogBB (-3-1.2) Predicted brain/blood partition coefficient; QPPMDCK (nm/s) <25-poor >500- great Predicted apparent MDCK cell permeability in nm/s. MDCK cells are considered to be a good mimic for the blood-brain barrier. QPlogKP- Predicted skin permeability; Q P log Khsa; Prediction of binding to human serum albumin; (acceptable range: -1.5 to 1.5); Percentage of human oral absorption; (<25% is poor and >80% is high).
Table 4: Evaluation of drug-like properties of the lead molecules by Qikprop Maestro.
TGFβ increases the tumor motility, invasion, and metastasis. Inhibition of TGFβ signaling offers a novel approach for the treatment of cancer. Monoclonal antibodies and small compounds used as a TGFβ inhibitor but due to cross activity these inhibitors fail in the preclinical trial. Our study showed some compounds has better docking energy such as EGCG and myricetin. Moreover, the RT-PCR showed that compound EGCG and myricetin reduced the mRNA expression of TGFβR1 at 80 μM concentration. This molecular docking study enhanced understanding of binding of compounds to the active site of proteins and to recapitulate the various binding energy, hydrophobic, hydrogen, an electrostatic bond that are important for the protein-ligand interactions. Further experimental work will be required for validation of our results.
6. Acknowledgement
We would like to thank Vice Chancellor, Central University of Punjab, Bathinda, Punjab, India for supporting this study with infrastructural requirements.
References
- Jakowlew SB. (2006). Transforming growth factor-ß in cancer and metastasis. Cancer and Metastasis Reviews. 25: 435-457.
- Ikushima H, Miyazono K. (2010). TGFß signalling: A complex web in cancer progression. Nature Reviews Cancer. 10: 415-424.
- van der Pluijm G. (2011). Epithelial plasticity, cancer stem cells and bone metastasis formation. Bone. 48: 37-43.
- Feng XH, Derynck R. (2005). Specificity and versatility in TGF-ß signaling through Smads. Annu Rev Cell Dev Biol. 21: 659-693.
- Kim SS, Shetty K, Katuri V, et al. (2006). TGF-ß signaling pathway inactivation and cell cycle deregulation in the development of gastric cancer: Role of the ß-spectrin, ELF. Biochemical and Biophysical Research Communications. 344: 1216-1223.
- Xu Y, Pasche B. (2007). TGF-ß signaling alterations and susceptibility to colorectal cancer. Human Molecular Genetics. 16: R14-R20.
- Yingling JM, Blanchard KL, Sawyer JS. (2004). Development of TGF-ß signalling inhibitors for cancer therapy. Nature Reviews Drug Discovery. 3: 1011-1022.
- Biswas S, Guix M, Rinehart C, et al. (2007). Inhibition of TGF-ß with neutralizing antibodies prevents radiation-induced acceleration of metastatic cancer progression. Journal of Clinical Investigation. 117: 1305.
- Hjelmeland MD, Hjelmeland AB, Sathornsumetee S, et al. (2004). SB-431542, a small molecule transforming growth factor-ß-receptor antagonist, inhibits human glioma cell line proliferation and motility. Molecular Cancer Therapeutics. 3: 737-745.
- Goel A, Kunnumakkara AB, Aggarwal BB. (2008). Curcumin as “Curecumin”: from kitchen to clinic. Biochemical Pharmacology. 75: 787-809.
- Singh P, Bast F. (2015). High-throughput virtual screening, identification and in vitro biological evaluation of novel inhibitors of signal transducer and activator of transcription 3. Medicinal Chemistry Research. 1-15.
- Singh P, Bast F. (2014a). In silico molecular docking study of natural compounds on wild and mutated epidermal growth factor receptor. Medicinal Chemistry Research. 1-12.
- Singh P, Bast F. (2014b). Multi-targeted molecular docking study of plant-derived natural products on phosphoinositide-3 kinase pathway components. Medicinal Chemistry Research. 23: 1690-1700.
- Singh P, Bast F. (2015a). Screening and biological evaluation of myricetin as a multiple target inhibitor insulin, epidermal growth factor, and androgen receptor; in silico and in vitro. Investigational New Drugs. 33: 575-593.
- Singh P, Bast F. (2015b). Screening of multi-targeted natural compounds for receptor tyrosine kinases inhibitors and biological evaluation on cancer cell lines, in silico and in vitro. Medical Oncology. 32: 1-18.
- Singh P, Singh RS, Rani A, Bast F. (2016). Homology modeling of chemokine CCR7, molecular docking and in vitro studies evidenced plausible immunotherapeutic anticancer natural compounds. Medicinal Chemistry Research. 1-15.
- Bhanot A, Sharma R, Noolvi MN. (2011). Natural sources as potential anti-cancer agents: A review. International Journal of Phytomedicine. 3: 9-26.
- Cho JY, Park J. (2008). Contribution of natural inhibitors to the understanding of the PI3K/PDK1/PKB pathway in the insulin-mediated intracellular signaling cascade. International Journal of Molecular Sciences. 9: 2217-2230.
- Cragg GM, Newman DJ. (2005). Plants as a source of anti-cancer agents. Journal of Ethnopharmacology. 100: 72-79.
- da Rocha AB, Lopes RM, Schwartsmann G. (2001). Natural products in anticancer therapy. Current Opinion in Pharmacology. 1: 364-369.
- Hillman GG. (2012). Dietary agents in cancer chemoprevention and treatment. Journal of Oncology. 1-2.
- Huang W, He T, Chai C, et al. (2012). Triptolide inhibits the proliferation of prostate cancer cells and down-regulates SUMO-specific protease 1 expression. Public Library of Science One. 7: e37693.
- Mayer AM, Gustafson KR. (2004). Marine pharmacology in 2001–2: Anti-tumour and cytotoxic compounds. European Journal of Cancer. 40: 2676-2704.
- Mayer AM, Gustafson KR. (2006). Marine pharmacology in 2003–2004: Anti-tumour and cytotoxic compounds. European Journal of Cancer. 42: 2241-2270.
- Mayer AM, Gustafson KR. (2008). Marine pharmacology in 2005–2006: Anti-tumour and cytotoxic compounds. European Journal of Cancer. 44: 2357-2387.
- Phosrithong N, Ungwitayatorn J. (2010). Molecular docking study on anticancer activity of plant-derived natural products. Medicinal Chemistry Research. 19: 817-835.
- Roell D, Baniahmad A. (2011). The natural compounds atraric acid and N-butylbenzene-sulfonamide as antagonists of the human androgen receptor and inhibitors of prostate cancer cell growth. Molecular and Cellular Endocrinology. 332: 1-8.
- Sarkar FH, Li Y. (2006). Using chemopreventive agents to enhance the efficacy of cancer therapy. Cancer Research. 66: 3347-3350.
- Sunil H. (2012). Inhibition studies of naturally occurring terpene based compounds with cyclin-dependent kinase 2 enzyme. Journal of Computer Science & Systems Biology. 1-2.
- Jorgensen WL, Duffy EM. (2002). Prediction of drug solubility from structure. Advanced Drug Delivery Reviews. 54: 355-366.
- Jorgensen WL, Maxwell DS, Tirado-Rives J. (1996). Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. Journal of American Chemical Society. 118: 11225-11236.
- Jorgensen WL, Tirado-Rives J. (1988). The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. Journal of American Chemical Society. 110: 1657-1666.
- Friesner RA, Banks JL, Murphy RB, et al. (2004). Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of Medicinal Chemistry. 47: 1739-1749.
- Friesner RA, Murphy RB, Repasky MP, et al. (2006). Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. Journal of Medicinal Chemistry. 49: 6177-6196.
- Halgren TA, Murphy RB, Friesner RA, et al. (2004). Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. Journal of Medicinal Chemistry. 47: 1750-1759.
- Lu JJ, Crimin K, Goodwin JT, et al. (2004). Influence of molecular flexibility and polar surface area metrics on oral bioavailability in the rat. Journal of Medicinal Chemistry. 47: 6104-6107.
- Liu LC, Tsao TCY, Hsu SR, et al. (2012). EGCG inhibits transforming growth factor-ß-mediated epithelial-to-mesenchymal transition via the inhibition of Smad2 and Erk1/2 signaling pathways in non-small cell lung cancer cells. Journal of Agricultural and Food Chemistry. 60: 9863-9873.
- Hayashi K, Takai S, Matsushima Nishiwaki R, et al. (2008). Epigallocatechin gallate reduces transforming growth factor ß-stimulated HSP27 induction through the suppression of stress-activated protein kinase/c-Jun N-terminal kinase in osteoblasts. Life Sciences. 82: 1012-1017.
- Tabuchi M, Hayakawa S, Honda E, et al. (2013). Epigallocatechin-3-gallate suppresses transforming growth factor-beta signaling by interacting with the transforming growth factor-beta type II receptor. World. 3: 100-107.
- Domitrovic R, Rashed K, Cvijanovic O, et al. (2015). Myricitrin exhibits antioxidant, anti-inflammatory and anti-fibrotic activity in carbon tetrachloride-intoxicated mice. Chemico-Biological Interactions. 230: 21-29.
- Kandasamy N, Ashokkumar N. (2014). Renoprotective effect of myricetin restrains dyslipidemia and renal mesangial cell proliferation by the suppression of sterol regulatory element binding proteins in an experimental model of diabetic nephropathy. European Journal of Pharmacology. 743: 53-62.
- Liu HL, Jiang WB, Xie MX. (2010). Flavonoids: recent advances as anticancer drugs. Recent Patents on Anti-Cancer Drug Discovery. 5: 152-164.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences