Genetic Diversity and Classification of Wild Arum (Araceae)Species Using Morphological Characters in Iran
Leila Joudi, Iraj Mehregan, Mostafa Assadi, Davoud Farajzadeh
Leila Joudi1, Iraj Mehregan1,*, Mostafa Assadi2, Davoud Farajzadeh3
1Department of Biology, Science and Research Branch, Islamic Azad University, Tehran, Iran;
2Research Institute of Forest and Ranglands, National Botanical Garden of Iran, Tehran, Iran;
3Department of Cellular and Molecular Biology, Faculty of Biological Sciences, Azarbaijan Shahid Madani University, Tabriz, Iran.
Received Date: May 24, 2016; Accepted Date: June 24, 2016; Published Date: July 01, 2016
Citation: Joudi L, Mehregan I, Assadi M, et al. Genetic diversity and classification of wild Arum (Araceae) Species Using Morphological Characters in Iran. Electronic J Biol, 12:3
Abstract
Assessment of genetic diversity is primarily useful to classification of plant species. In this study, a multivariate statistical analysis was performed on morphological characters of Arum L. species in Iran. Totally, 29 qualitative and quantitative morphological characters of species were evaluated. Cluster analyses by ward method classified the genotypes based on morphological traits in three groups. Cluster 1 had 1 genotype. Cluster 2 had 2 genotypes and cluster 3 had 2 groups. Dendrogram of Genetic Relationships among Arum species which was constructed by the UPGMA method demonstrated that 4 main groups existed in the collection at the highest level of hierarchy (0.34). Using the results from morphological studies, differential characteristics were obtained in the principal component analysis. The result of principal component analyses introduced two principal components with eigenvalue more than one which contributed 99.075% of the total variability. Classification of the cluster analysis was confirmed by principal component analysis and good variety of different traits to species based on similarities and differences have separated. In conclusion, morphological traits can identify and classify species of this genus as a systematic application.
Keywords
Arum; Cluster analysis; Principal component analysis.
Introduction
The family Araceae with 3790 species in 117 genera, have a worldwide distribution and are found in a wide range of environments [1,2]. Most of the aroids are tropical and contain members from terrestrial, aquatic, and epiphytic habitats although there are many aroids indigenous to temperate climates. The family Araceae is most easily diagnosed by the inflorescence which a spadix unbranched spike is bearing small bractless flowers subtended by a modified leaf called a spathe. The plants have adapted in response to climatic, ecological and biotic conditions (i.e., selective pressures) according to the various habitats occupied in the different regions of Europe and the Middle East [3]. The genus Arum with 28 species have distributed in Europe, North Africa, the Middle East and Central Asia [4,5]. In Iran, Arum species grow naturally in mountains, red and alluvial soils, near water canal, rocky places and forests. Amorphophallus Decne. is especially noteworthy because of the enormous tubers produced by certain species (Arum giganteum). The largest species: Amorphophallus titanum Becc. can produce tubers weighing approximately 70 kg. Genetic variability is more likely to be found between species from completely different environmental conditions [6]. Evidence from morphology, as an important feature in the diagnosis and taxonomy of plants and as a basic language used to describe and identify, classify and study the relationship between evolutionary and phylogenetic relationships of plants, through morphological characteristics has always been popular [7]. The aim of this study was to evaluate several quantitative and qualitative characteristics of the species, with emphasis on the traits of spath and spadix which could be used as important characteristics to distinguish the species of this genus for taxonomic identification.
Materials and Method
Plant material and morphological evaluation
In order to conduct morphometric researches, all species of Arum were collected from different locations of Iran reported in the flora of Iran that Arum species were introduced before [8]. The number of collected plants in each location was 5 plants. The collected plants had an uneven distribution in different parts of Iran: A. maculatum from 11 locations. (North, North West), A. kotschyi from 12 locations (North, North West, North East, Tehran), A. korolkowii from 11 locations (North, North West, North East, Tehran), A. virescens from 9 locations (North, North West, East), A. conophalloides from 8 locations (East), A. giganteum from 3 locations (East and Center). Table 1 represents only the samples which already have a herbarium code. The selected plants were growing under ambient environmental conditions like e.g. light, temperature, soil condition, etc. The minimum and maximum of each parameter were recorded. Due to the small differences in between species, the values would overlap if the average has taken into account. The samples were sent to the Forests and Rangelands Institute (TARI) and were identified based on Flora of Iran and the world scientific resources [8]. Lab analyses were carried out at the Herbarium of Research Institute of Forests and Rangelands (TARI) in 2014 and 2015. The following quantitative morphological variable were petiole length (cm), leaf blade length and wide (cm), peduncle (cm), spathe length and wide (cm), spathe tube length (cm), pistillate zone, staminode zone and staminate zone (mm). Some of morphological characters were not measurable quantitatively and qualitative characters were used for analysis. Qualitative characters used in the experiment were as follows: Tuber shape, arrangement of leaves and inflorescence, leaf blade shape, leaf blade apex and inner and outer section of spath. In order to conduct cluster analysis, the minimum and maximum yield was used, while qualitative characteristics were coded as attributes of two or more cases.
| Species | Origin, voucher |
|---|---|
| Arum maculatum | Iran, Prov. N. Gorgan, Ramsar 200-500 m, (69216 TARI). |
| A. kotschyi | Iran, Prov. N. Mazandaran, Siahbishe 2200 m, (27355 TARI). |
| A. korolkowii | Iran, Prov. N. Mazandaran, veisar 1600 m, (50656TARI). |
| A. virescens | Iran, Prov. N. Mazandaran, Polsefid 670 m, (36826TARI). |
| A. conophalloides | Iran, Prov. W. Hamedan, Nahavand 2600 m, (22222TARI). |
| A. giganteum | Iran, Prov. W. Khorramabad, Rimele 1800 m, (2558TARI). |
Table 1. Species characteristic and species herbarium code.
Statistical analysis
Collected data in 2014 and 2015 years were combined and analyzed. Statistical analysis and draw dendrograms were done by SAS 9.1. The mean and standard deviation was prepared and shown in Table 2. The correlation analyses were performed following the methods of and Snedecor and [9,10]. In order to describe the classification of studied species cluster analysis using the ward and principal component analysis (Factor Analysis) and also ordination attributes (Principal Component Analysis) were performed on traits. The Euclidean distance coefficients were used in the assessment of the similarity of morphological characters in the cluster analysis Table 3 [11].
| Variables | N | Mean | Std. Deviation |
|---|---|---|---|
| Petiole length(min) | 6 | 17.8333 | 9.82683 |
| Petiole length (max) | 6 | 40.6667 | 8.43010 |
| Leaf Blade Length ( min) | 6 | 11.8333 | 8.93122 |
| Leaf Blade Length (max) | 6 | 20.0000 | 5.32917 |
| Leaf Blade Wide (min) | 6.0000 | 4.64758 | |
| Leaf Blade Wide (max) | 6 | 11.6667 | 2.50333 |
| Peduncle Length (min) | 6 | 20.5000 | 12.40564 |
| Peduncle Length (max) | 6 | 44.6667 | 15.97081 |
| Spathe Length (min) | 6 | 17.1667 | 12.96791 |
| Spathe Length (max) | 6 | 32.0000 | 20.27807 |
| Spathe Wide (min) | 6 | 3.6667 | 2.42212 |
| Spathe Wide (max) | 6 | 6.7500 | 4. 60163 |
| Spathe Tube Length (min) | 6 | 15.0000 | 12.74363 |
| Spathe Tube Length (max) | 6 | 25.8333 | 20.49797 |
| Spathe Tube Limb (min) | 6 | 11.1667 | 12.09408 |
| Spathe Tube Limb (max) | 6 | 20.9167 | 20.50467 |
| Pistillode Zone (min) | 6 | 13.5000 | 8.64292 |
| Pistillode Zone (max) | 6 | 21.6667 | 11.25463 |
| Staminate Zone (min) | 6 | 6.3333 | 4.45720 |
| Staminate Zone (max) | 6 | 8.5000 | 4.23084 |
| Staminode Zone (min) | 6 | 6.1667 | 4.91596 |
| Staminode Zone (max) | 6 | 9.5000 | 4.84768 |
| Tuber Shape | 6 | 1.1667 | 0.40825 |
| Leaf O Flower | 6 | 1.8333 | 0.40825 |
| Leaf Blade Shape | 6 | 1.5000 | 0.83666 |
| Leaf Blade Apex | 6 | 1.1667 | 0.40825 |
| Spathe Tube Color | 6 | 1.5000 | 0.83666 |
| Inner Section of Spathe | 6 | 2.1667 | 0.75277 |
| Outer Section of Spathe | 6 | 1.8333 | 0.40825 |
| Valid N (listwise) | 6 |
Table 2. Mean and standard deviation of each variable.
| Species | Petiole length (cm) |
Leaf Blade Length (cm) |
Leaf Blade Wide (cm) | Peduncle Length (cm) | Spathe Length (cm) | Spathe Wide (cm) | Spathe Tube Length (cm) | Spathe Tube Limb (cm) | Pistillode Zone (mm) | Staminate Zone (mm) | Staminode Zone (mm) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Arum maculatum | 10-74 | 7-22 | 2-10 | 4-18 | 11-19 | 4.5-7.5 | 6-9 | 3-6.5 | 6-10 | 2-3 | 3-4 |
| Arum kotschyi | 9 -37 | 8-17 | 3-13 | 13-51 | 7.5-17 | 1.5-3 | 6-13 | 4-10 | 7-15 | 3-5 | 3-7 |
| Arum korolkowi | 12-35 | 8-16 | 5-13 | 16-64 | 14-20 | 1.5-2 | 11-17 | 5-6 | 12-15 | 6-8 | 5-7 |
| Arum virescens | 21-55 | 9-17 | 5-8 | 35-55 | 13-27 | 3.5-6 | 12-21 | 8.5-18 | 13-20 | 5-10 | 5-8 |
| Arum conophalloides | 20-35 | 9-18 | 6-11 | 20-40 | 15-39 | 3-7 | 15-30 | 11.5-25 | 13-40 | 6-10 | 5-15 |
| Arum giganteum | 15-35 | 10-30 | 10-15 | 35-40 | 43-70 | 8-15 | 40-65 | 35-60 | 20-30 | 10-15 | 6-16 |
Table 3. Morphological quantitative variable in species characteristic.
Results
Correlation
The correlations among all pairs of variables are shown in Table 4. Petiole length (min) was significantly correlated with leaf blade length (r=0.891*), leaf blade wide (r=0.942**) and spath length (r=0.960**). On the other hand spath length could be effected on spath tube length (r=0.990**), spath tube limb (r=0.986**), pistillode zone (r=0.957**), staminate zone (r=0.954**) and staminode zone (r=0.951**). Staminate zone was correlated on pistillode zone(r=0.992**), Staminode zone effected on pistillode zone(r=0.986**) and staminate zone (0.992**). In qualitative traits, arrangement of leaves and flowers was effective on tuber shape (1.000**). The other characters expressed a non-significant correlation. Therefore, the function of the each of the traits is assessed in its performance on petiole length and inflorescence components. Positive and significant correlation between petiole length and the structure of florescence could be originated from genotypes ability in competition of light absorption in order to promote the photosynthesis process to reach to the reproductive stage.
| PL min |
PL max | LBL min | LBL max |
LBW min |
LBW max |
PEL min |
PEL max |
SL min |
SL max |
SW min |
SW max |
STL min |
STL max |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PLmin | ||||||||||||||
| PL max | -0.117 | |||||||||||||
| LBL min | 0.891* | -0.325 | ||||||||||||
| LBL max | 0.733 | -0.187 | 0.895* | |||||||||||
| LBW min | 0.942** | -0.378 | 0.968** | 0.791 | ||||||||||
| LBW max | 0.306 | -0.859* | 0.632 | 0.495 | 0.602 | |||||||||
| PEL min | 0.846* | 0.147 | 0.631 | 0.333 | 0.725 | 0.045 | ||||||||
| PEL max | -0.022 | -0.15 | -0.099 | -0.512 | 0.043 | 0.142 | 0.394 | |||||||
| SL min | 0.914* | -0.325 | 0.981** | 0.886* | 0.979** | 0.587 | 0.631 | -0.111 | ||||||
| SL max | .960** | -.335 | .940** | 0.829* | 0.968** | 0.496 | .681 | -0.149 | 0.957** | |||||
| SW min | 0.821* | 0.023 | 0.866* | 0.961** | 0.791 | 0.275 | 0.486 | -0.500 | 0.878* | 0.849* | ||||
| SW max | 0.861* | -0.070 | 0.877* | 0.946** | 0.823* | 0.304 | 0.511 | -0.498 | 0.886* | 0.906* | 0.982** | |||
| STL min | 0.947** | -0.346 | 0.977** | 0.836* | 0.996** | 0.577 | 0.700 | -0.041 | 0.990** | 0.979** | 0.839* | 0.868* | ||
| STL max | 0.956** | -0.375 | 0.959** | 0.802 | 0.991** | 0.564 | 0.715 | -0.034 | 0.967** | 0.991** | 0.808 | 0.859* | 0.992** |
Table 4: Correlation coefficients between studied traits expressed as averages combining two years. (*p<0.05, ** p<0.01).
| STM min |
STM max |
PZ min |
PZ max |
STZ min |
STZ max |
SDZ min |
SDZ max |
TS | LOF | LBS | LBA | STC | ISS | OSS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PL min | |||||||||||||||
| PL max | |||||||||||||||
| LBL min | |||||||||||||||
| LBL max | |||||||||||||||
| LBW min | |||||||||||||||
| LBW max | |||||||||||||||
| PEL min | |||||||||||||||
| PEL max | |||||||||||||||
| SL min | |||||||||||||||
| SL max | |||||||||||||||
| SW min | |||||||||||||||
| SW max | |||||||||||||||
| STL min | |||||||||||||||
| STL max |
| PL min |
PL max |
LBL min |
LBL max |
LBW min |
LBW max |
PEL min |
PEL max |
SL min |
SL max |
SW min |
SW max |
STL min |
STL max |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| STM min | 0.950** | -0.325 | 0.982** | 0.855* | 0.987** | 0.560 | 0.697 | -0.090 | 0.980** | 0.986** | 0.857* | 0.894* | 0.995** | 0.994** |
| STM max | 0.960** | -0.293 | 0.956** | 0.833* | 0.965** | 0.498 | 0.715 | -0.125 | 0.948** | 0.990** | 0.850* | 0.904* | 0.975** | 0.988** |
| PZ min | 0.955** | -0.310 | 0.957** | 0.773 | 0.996** | 0.545 | 0.768 | 0.072 | 0.975** | 0.957** | 0.793 | 0.816* | 0.991** | 0.982** |
| PZ max | 0.663 | -0.393 | 0.421 | 0.250 | 0.574 | 0.166 | 0.516 | -0.002 | 0.47 | 0.688 | 0.318 | 0.473 | 0.565 | 0.652 |
| STZ min | 0.919** | -0.390 | 0.966** | 0.808 | 0.994** | 0.621 | 0.680 | 0.024 | 0.988** | 0.954** | 0.800 | 0.819* | 0.993** | 0.977** |
| STZ max | 0.945** | -0.241 | 0.802 | 0.523 | 0.915* | 0.359 | 0.894* | 0.275 | 0.837* | 0.886* | 0.605 | 0.660 | 0.894* | 0.912* |
| SDZ min | 0.920** | -0.327 | 0.989** | 0.855* | 0.989** | 0.607 | 0.680 | -0.027 | 0.994** | 0.951** | 0.845* | 0.855* | 0.993** | 0.973** |
| SDZ max | 0.833* | -0.494 | 0.704 | 0.503 | 0.817* | 0.429 | 0.634 | 0.028 | 0.730 | 0.879* | 0.528 | 0.657 | 0.806 | 0.868* |
| TS | -0.391 | 0.368 | -0.265 | 0.184 | -0.422 | -0.326 | -0.652 | -0.818* | -0.233 | -0.314 | 0.169 | 0.080 | -0.346 | -0.402 |
| LOF | 0.391 | -0.368 | 0.265 | -0.184 | 0.422 | 0.326 | 0.652 | 0.818* | 0.233 | 0.314 | -0.169 | -0.080 | 0.346 | 0.402 |
| LBS | -0.499 | -0.425 | -0.308 | -0.493 | -0.257 | 0.382 | -0.318 | 0.674 | -0.304 | -0.460 | -0.642 | -0.688 | -0.319 | -0.356 |
| LBA | 0.108 | -0.329 | -0.155 | -0.184 | 0.000 | -.0130 | -0.020 | -0.143 | -0.082 | 0.169 | -0.135 | 0.027 | 0.000 | 0.100 |
| STC | -0.304 | 0.766 | -0.335 | 0.045 | -0.463 | -0.668 | -0.356 | -0.644 | -0.082 | -0.365 | 0.148 | 0.039 | -0.394 | -0.449 |
| ISS | 0.734 | -0.557 | 0.600 | 0.299 | 0.743 | 0.460 | 0.653 | 0.288 | 0.611 | 0.760 | 0.311 | 0.447 | 0.709 | 0.780 |
| OSS | 0.391 | -0.368 | 0.265 | -0.184 | 0.422 | 0.326 | 0.652 | 0.818* | 0.233 | 0.314 | -0.169 | -0.080 | 0.346 | 0.402 |
| STM min |
STM max |
PZ min |
PZ max |
STZ min |
STZ max |
SDZ min |
SDZ max |
TS | LOF | LBS | LBA | STC | ISS | OSS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| STM min | |||||||||||||||
| STM max | 0.991** | ||||||||||||||
| PZ min | 0.979** | 0.954** | |||||||||||||
| PZ max | 0.578 | 0.646 | 0.555 | ||||||||||||
| STZ min | 0.976** | 0.942** | 0.992** | 0.525 | |||||||||||
| STZ max | 0.878* | 0.878* | 0.933** | 0.714 | 0.891* | ||||||||||
| SDZ min | 0.985** | 0.954** | 0.986** | 0.464 | 0.992** | 0.861* | |||||||||
| SDZ max | 0.815* | 0.855* | 0.795 | 0.935** | 0.778 | 0.863* | 0.734 | ||||||||
| TS | -0.331 | -0.344 | -0.425 | -0.508 | -0.366 | -0.637 | -0.316 | -0.556 | |||||||
| LOF | 0.331 | 0.344 | 0.425 | 0.508 | 0.366 | 0.637 | 0.316 | 0.556 | 1.000** | ||||||
| LBS | -0.385 | -0.475 | -0.263 | -0.425 | -0.215 | -0.254 | -0.267 | -0.370 | -0.293 | ||||||
| LBA | 0.014 | 0.098 | -0.028 | 0.798 | -0.037 | 0.174 | -0.116 | 0.556 | -0.200 | -0.293 | |||||
| STC | -0.376 | -0.370 | -0.429 | -0.531 | -0.429 | -0.537 | -0.365 | -0.616 | 0.878* | -0.429 | -0.293 | ||||
| ISS | 0.710 | 0.746 | 0.722 | 0.905* | 0.695 | 0.848* | 0.640 | 0.959** | -0.759 | -0.159 | 0.542 | -0.794 | |||
| OSS | 0.331 | 0.344 | 0.425 | 0.508 | 0.366 | 0.637 | 0.316 | 0.556 | -1.000** | 1.000** | 0.293 | 0.200 | -0.878* | 0.759 |
Cluster analyses
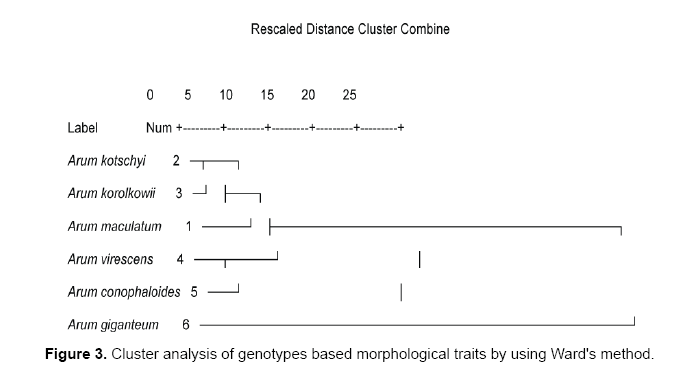
Data analyses using Ward and UPGMA methods are shown in Figures 1 and 2. Cluster analyses by ward method based on all morphological characters from each genotype shown in Figure 3. Cluster analysis categorized genotypes into three groups. Cluster 1 had 1 genotype. Cluster 2 had 2 genotypes and cluster 3 had 2 groups. Cluster 3 could be further divided into 2 subgroups. The first cluster is allocated only Arum giganteum that is endemic in west Iran and has its own unique morphological characteristics. Cluster 2 contained 2 species including Arum conophalloides and Arum viresences. Cluster 3 was divided into 2 groupings, including Arum maculatum and sub groups with Arum kotschyi and Arum korolkowii.
Dendrogram of genetic relationships among Arum species constructed by the UPGMA method demonstrated that 4 main groups existed at the highest level of hierarchy (0.34). According to the results of dendrogram A. giganteum and A. maculatum replaced in independent groups. Group 3 divided into 2 subgroups consist of A. conophalloides and subgroup 2. The second subgroup included A. virescense, A.kotschyi and A. korolkowii.
Principal components analysis (PCA)
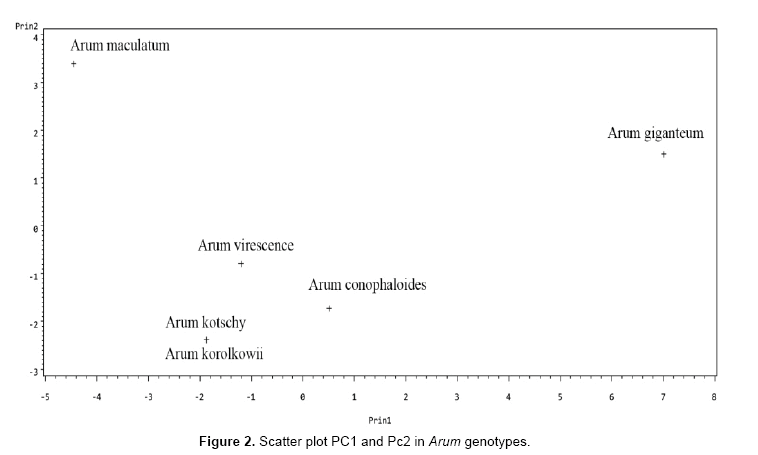
The results of principal component analysis on Arum species data, including the Eigen values and cumulative variances of correlation matrix of Arum species traits are presented in Tables 5 and 6. It is clear that the first principal component of Arum species data accounts 62.256% of total variability in the data, while the second principal component accounts for 20.680% of the total variability. Finally, together the first and second components accounts for 82.936% of the total variability.
| Principal component | Total | % of Variance | Cumulative % |
|---|---|---|---|
| PC1 | 18.054 | 62.256 | 62.256 |
| PC2 | 5.997 | 20.680 | 82.936 |
| PC3 | 2.572 | 8.868 | 91.804 |
| PC4 | 2.109 | 7.271 | 99.075 |
Table 5: Eigen value, proportion and cumulative explained by two PC Principal Component Eigen value proportion cumulative.
| Traits | PC1 | PC 2 | Traits | PC1 | PC 2 |
|---|---|---|---|---|---|
| PL(min) | 0.959* | 0.110 | STM max | 0.982* | 0.145 |
| PL max | -0.379 | 0.426 | PZ min | 0.982* | 0.040 |
| LBL min | 0.944* | 0.205 | PZ max | 0.676 | -0.271 |
| LBL max | 0.760 | 0.598* | STZ min | 0.971* | 0.072 |
| LBW min | 0.989* | 0.035 | STZ max | 0.934* | -0.203 |
| LBW max | 0.555 | -0.213 | SDZ min | 0.963* | 0.148 |
| PEL min | 0.747 | -0.204 | SDZ max | 0.885 | -0.209 |
| PEL max | 0.025 | -0.874 | TS | -0.462 | 0.854 |
| SL min | 0.952* | 0.220 | LOF | 0.462 | -0.854 |
| SL max | 0.984* | 0.151 | LBS | -0.334 | -0.616 |
| SW min | 0.772 | 0.631* | LBA | 0.120 | -0.274 |
| SW max | 0.829 | 0.548* | STC | -0.492 | 0.815* |
| STL min | 0.985* | 0.119 | ISS | 0.813 | -0.473 |
| STL max | 0.998* | 0.062 | OSS | 0.462 | -0.854 |
| STM min | 0.986* | 0.149 |
Table 6: Vector loadings explained by the first two PC.
The first four principal components are orthogonal with each other and extract maximum of total variability (about 99.075%). Based on the correlation between features and components are given in Table 5: In first principal component petiole long, leaf blade length and wide, spath long, spath tube long, staminode, staminate and pistillode zone have positive correlation. The second principal component; spath wide and spath tube color had positive correlation.
Discussion
There are six species of Arum in Iran that most of them have been adapted in north and west. In the current research, Arum species from different places were analyzed using morphological characters using Ward, UPGMA and PCA methods. Classical taxon definition and circumscription in the genus Arum only partially match our morphometrictraits [12]. As shown in Figures 1 and 3, identity the sections and subsections and species are extremely challenged and it seems obvious that there are some varieties in classification of plants in Iran [5]. In the research conducted by Peter Boyce in 1993, Arum rupicola and Arum conophalloides reported as synonym plants. In Turkey and Iran, Lance Chilton discovered the A. rupicola on the Aegean island of Lesbos which usually displays a massive conic-clyindric spadix-appendix borne on a short, rather stout stipe; this form is the plant known as A. conophalloides. Toward the Turkish border with Syria, the spadix appendix is often much more slender with a rather poorly defined stipe, this is the plant described as A.rupicola. A. rupicola in other classification of Boyce and has been situated in Tenufila subsection with A. jacquemonti and A. korolkowii. Based on morphological characteristics and using the Ward and UPGMA analyses A. korolkowii is completely separated from A. conophalloides and A. giganteum and were introduced as independent plants.
Engler's taxon contains 8 species that A. italicum and A. maculatum differ markedly because of their horizontal-rhizomatous tubers. In this study, A. maculatum replaced in separated group. A. conophalloides is a closer match to the A. protologue than A. jacquemontii. The only change to be made is the adoption of Boissier's earlier name, A. rupicola, for the species.
In another study the color of floral chamber wall was used as a feature to help the classification. The floral chamber wall can be seen bicoloured: dark purple in its upper part and pale green (translucent) in its lower part. This kind of floral chamber wall is observed in A. orientale, A. rupicola, A. purpureospathum, and A. elongatum, but not in A. maculatum, A. italicum, A. concinnatum or A. cylindraceum. This character also can separate A. maculatum and A. kotschyi and A. korolkowii that can be placed with A. rupicola in the same groups. Also regarded to Boyce, ecologically, differences between two types of cryptic and flag inflorescence can separated A. maculatum and A. korolkowii whereas cryptic inflorescence species like A. maculatum grow in wooded or scubby area, but flag species (A. korolkowii and A. rupicola) usually inhabit in open or rocky areas.
The results of PCA showed peduncle length aligned with the characteristics of inflorescences. Field studies suggest that peduncle length can be an important factor in pollination biology and investigation by has shown that spadix appendix odour is of considerable importance in determining the predominant pollinator plants [13]. Bedalov and Grayum worked on pollen of some and noted the similarity in the pollen of different species [14,15]. It will be interesting to see which separates the morphological characteristics of some species and can also be used to separate the species Iran by the correlation characteristic between species. Makhadmeh et al. showed that Arum populations of the same species or having a common genome were grouped in the same cluster, regardless of the collection site [16,17]. The wide range of genetic distance was represented by the high level of DNA polymorphism occurring among Arum species.
Conclusion
The current study assessed the levels of genetic variation of 6 Arum species in Iran, to provide a baseline for further studies and conservation strategies. Molecular markers significantly need for separation between Arum species. Also based on this study, a chemo diversity study of these species is recommended, as well as the establishment of in situ and ex situ field gene banks to protect this plant and the development of legal measures to conserve this species and consider this plant as one of those that must be protected.
Acknowledgement
The authors would like to express their thanks to Islamic Azad University, Tehran, Science and Technology Research Branch for providing the facilities necessary to carry out the work.
References
- Boyce PC, Croat TB. (2011).Theuberlist of Araceae, totals for published and estimated number of species in aroid genera. 2-3.
- Espindola A, Buerki S, Bedalov M, Kupfer PH, Alvarez N. (2010). New insights into the phylogenetics and biogeography of Arum (Araceae): Unravellingits evolutionary history. Bot J Linn Soc. 163:14–32.
- Gibernau M, Macquart D, Przetak G. (2004). Pollination in the genus Arum: A review. Aroideana.27:148-166.
- Mayo SJ, Bogner J, Boyce PC. (1997).The Genera of Araceae. Royal Botanical Gardens, Kew. 370.
- Boyce PC. (1989). A new classification of Arum with keys to the infrageneric taxa. Kew Bulletin.44:383–395.
- Gustin DL, Huff DR. (1999). Genetic variation within and among white clover populations from managed permanent pastures of the northeastern USA. Crop Sci.39: 524-530.
- Stace CA. (1991). Plant taxonomy and biosystematics. Cambridge University Press. 71-73.
- Assadi M (1988). Flora of Iran.Research Institute of Forest and Rangelands, National Botanical Garden of Iran.2:30.
- Dewey DR, Lu KH. (1959). A correlation and path coefficient analysis of components of crested wheat grass and seed production. Agron J. 51:515-518.
- Snedecor W, Cochran WG. (1989). Statistical methods. Oxford and IBM Calcutta. 593.
- Naroui Rad MR, Farzanju M, Fanay HR, PolshekanePahlevan MR. (2006). The study genetic variation and factor analysis for morphological characters of wheat native accessions of Sistan and Baluchistan. Pajouhesh&Sazandegi.73:50-57.
- Boyce PC. (2006).Arum – a decade of change. Aroideana.29:132–139.
- Koach J. (1987). Bio-ecological studies of flowering and pollination in Israeli Araceae. PhD thesis, University of Tel-Aviv.
- Bedalov M.(1985). Scanning electron microscopy of pollen grains of some species of the genus Arum (Araceae). Pl SystEvol. 149:211-216.
- Grayum MH. (1984). Palynology and phylogeny of the Araceae. PhD thesis, University of Massachusetts.
- Makhadmeh I, Al-Lozi S, Duwayri M, Shibli RA, Migdadi H. (2010). Assessment of genetic variations in Wild Arumspecies from Jordan using amplified fragment length polymorphism (AFLP) Markers. JJAS 6:224-239.
- Engler A. (1920).Arum. Das Pflanzenr. 73:67-99.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences