Detection of Aneuploidy in Neural Stem Cells of the Developing and Adult Human Brain
Ivan Y. Iourov, Svetlana G. Vorsanova, Yuri B. Yurov
Ivan Y. Iourov1,2,*,Svetlana G. Vorsanova1,2,Yuri B. Yurov1,2
1National Research Center of Mental Health,Russian Academy of Medical Sciences,Moscow,119152,Russian Federation
2Institute of Pediatrics and Children Surgery,Rosmedtechnologii,Moscow,127412,Russian Federation.
Abstract
Neural stem cells (NSCs) are found to be present in the developing and adult human brain. The genuine interest to the properties of NSCs in the light of recent advances in neurosciences, developmental biology and molecular medicine has forced to reconsider genetic features of the developing and adult human brain, which are, as yet, incompletely known. A series of articles has reported that genomic variations manifested as aneuploidy (losses/gains of whole chromosomes in a cell) hallmarks the development of the human brain and that aneuploid cells populate the adult human brain. This has led to the creation of a new biomedical direction termed molecular neurocytogenetics. Although the studies in the field use previous developments of molecular cytogenetics, there is apparent interest in considering both advantages and disadvantages of the techniques in context of studying NSCs in the developing and adult human brain. The outcomes of such evaluation will be probably found valuable for designing future NSC studies. Here, molecular cytogenetic approaches allowing single-cell monitoring of chromosome complement variations are considered in context of different applications in NSC research. The data presented simplifies the choice between molecular cytogenetic techniques for obtaining better results during forthcoming NSC studies.
Keywords
aneuploidy; human brain; fluorescence in situ hybridization (FISH); molecular cytogenetics; neural stem cells (NSCs).
1. Introduction
Neural stem cells (NSCs) populate the developing human brain and are known to be involved in adult neurogenesis of human [1,2]. This assumes NCS genetic properties to be considered a determinant of the human brain development and functioning as during intrauterine period as during ontogeny,as a whole. In this context,the intercellular genomic variations that possess probably the most appreciable impact on brain development and neurogenesis are related to aneuploidy or gain/loss of whole chromosomes in a cell. This is due to simultaneous losses or gains of hundreds or thousands genes of a chromosome involved in aneuploidy. Affecting large proportions of neural cells,aneuploidy is usually devastative and is suggested to hallmark numerous pathogenic processes in the human brain. In contrast,smaller aneuploid cell populations are considered an integral part of the human brain contributing to neuronal diversity. [3]. Recent studies have shown the developing human brain to be populated by aneuploid neural cells [4,5]. The adult human brain contains significantly lower amount of aneuploid cells that is still appreciable [4,6,7]. The increase of aneuploidy rates in the adult human brain is shown to be a likely pathogenic mechanism for major psychiatric disorders (schizophrenia and autism) [8-10]. These results have put forward a new biomedical direction termed molecular neurocytogenetics that is defined as the survey of chromosome complement and behaviour in the human brain [3]. Together,this suggests that studying structural and functional genome organization in the human brain is promising and important area of biomedical research. Although some progress has been made towards technical developments in molecular cytogenetics of the human brain [3-13],numerous technological aspects are not completely worked out. The objectives of the present communication are to consider both advantages and disadvantages of advanced molecular cytogenetic techniques for single-cell monitoring of chromosome complement variations. Once presented,it will probably simplify the choice between molecular cytogenetic techniques for obtaining better results during molecular neurocytogenetic evaluations of NSCs alone or in combination with other cytochemical and histochemical techniques for NSC analysis.
2. Aneuploidy in the developing and adult human brain
It is commonly accepted that genetic features of cells populating the developing and adult human brain determines its development and functioning. NSCs are not an exception [1-3,14]. Aneuploidy is suggested to be among the most common types of intercellular genomic variations that have exceedingly appreciable effect on the human brain [3-13].
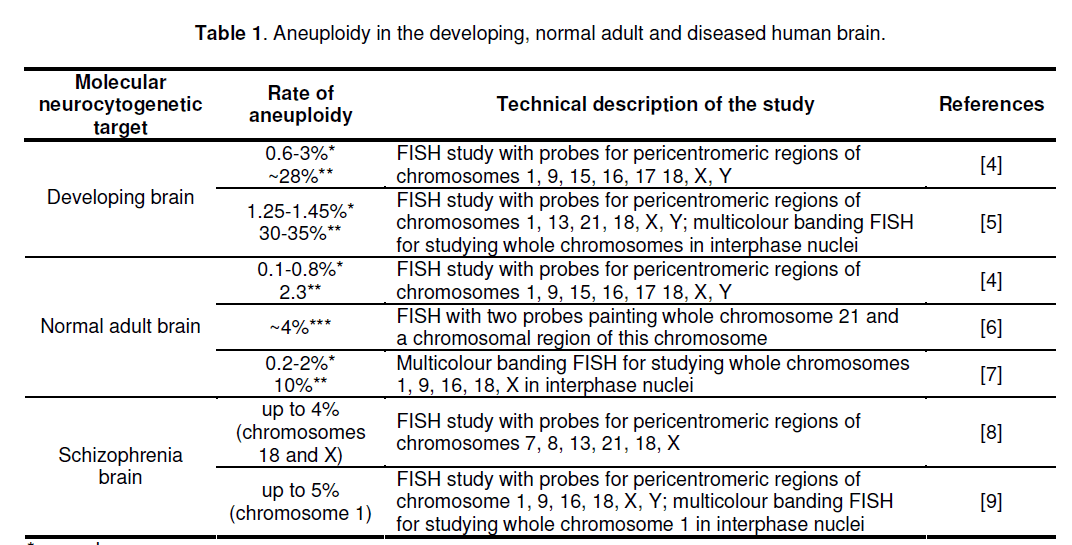
Until molecular cytogenetic techniques,allowing chromosome studying at single-cell level and at molecular resolutions,were developed,the definition of aneuploidy in the human brain was impossible. This was further complicated by impossibility to apply standard cytogenetic techniques (metaphase cytogenetics) for analysis of the human brain. The filling of this gap launched a series of studies that uncovered the frequencies of aneuploidy in the human brain. The most efficient techniques for the assessment of single-cell genomic variations in the human brain were found those based on fluorescent in situ hybridization (FISH). The latter allows visualization of either specific chromosome regions or whole chromosomes in their integrity (for review see [3,12]). Thus,the developing human brain was shown to possess exceedingly high rate of aneuploid cells [4,5,11]. The rate of aneuploidy in the adult human brain was found to be significantly lower as to the developing brain without direct evidences for changing through aging [4,6,7] (Table 1). However,there is a discrepancy of brain aneuploidy studies that is,in major part,produced by differences between techniques applied [3]. Nevertheless,molecular cytogenetic analyses of the diseased brain were able to depict that aneuploidy is a pathogenic mechanism in at least a number of schizophrenia cases [8,10]. Taking into account the problems that were reported during molecular neurocytogenetic studies [3-14],it is to conclude that technical side of molecular neurocytogenetics is highly important and should be critically addressed before a study is launched.
3. FISH and FISH-based techniques
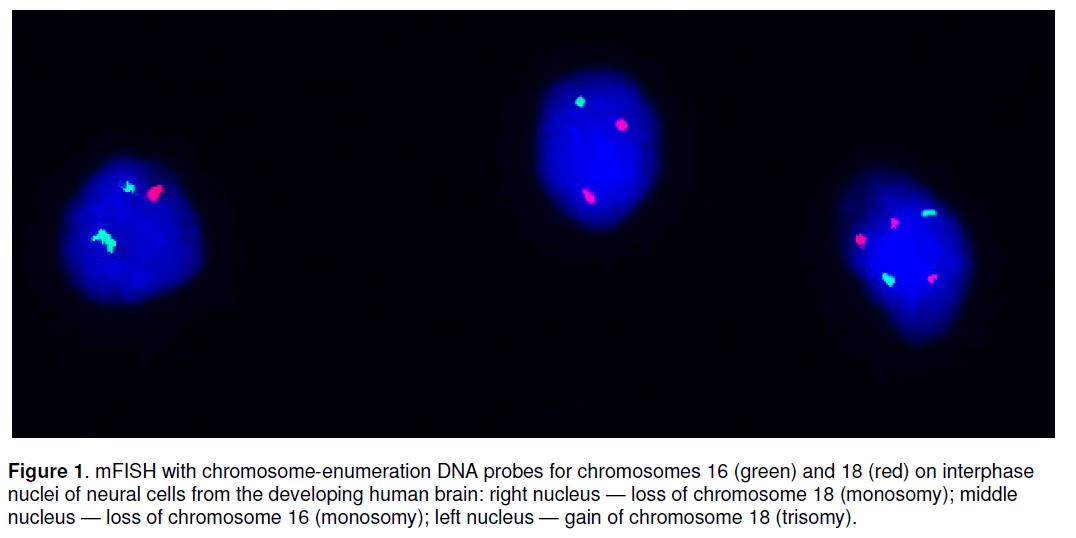
Current molecular cytogenetics possesses numerous approaches towards high-resolution analysis of interphase chromosomes. Among them,FISH-based techniques are considered as more efficient for studying aneuploidy in somatic cells,especially,in cases of aneuploidy affecting small cell populations [3,12]. FISH achieves better resolution when multicolour protocols are applied (for review see [15]). Multicolour FISH approaches are found highly applicable in diagnostic and basic biomedical research [15,16]. In context of studying aneuploidy in NSCs (i.e. single-cell analysis of large cell populations),FISH technique,that applies several chromosome-enumeration DNA probes each labelled by corresponding colour [17],appears to be of sufficient efficiency for such applications. This is further supported by studies of different human somatic cells (including NSCs) [3-5,9-12,16-18]. Until recently,multicolour FISH with chromosomeenumeration DNA probes (mFISH) was almost unique approach towards high-resolution single-cell molecular cytogenetic analysis of aneuploidy in interphase nuclei [3,4,12,16,18] and was found to be applicable of studying aneuploidy in the developing and adult human brain [3,4,8]. Figure 1 demonstrates an example of mFISH on interphase nuclei of neural cells from the developing human brain.
Figure 1. mFISH with chromosome-enumeration DNA probes for chromosomes 16 (green) and 18 (red) on interphase nuclei of neural cells from the developing human brain: right nucleus — loss of chromosome 18 (monosomy); middle nucleus — loss of chromosome 16 (monosomy); left nucleus — gain of chromosome 18 (trisomy).
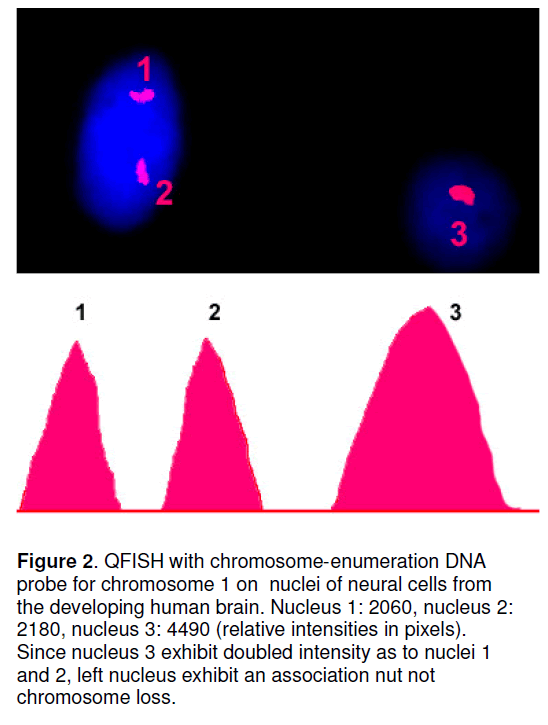
The efficiency of mFISH aneuploidy detection in human brain cells becomes higher since the introduction of quantitative FISH (QFISH) [19]. The results of simple mFISH approaches are seriously affected due to specific organization of interphase chromosomes in the human brain that is usually manifested as chromosomal associations [3,5,7]. In other terms,traditional mFISH techniques poorly differentiate between specificity of nuclear organization (for example,chromosome associations) and aneuploidy appearing as chromosomal loss or monosomy [19]. To solve this,one can apply additionally QFISH (Figure 2 ) that allows such differentiation and is shown to be highly efficient for aneuploidy detection in the human brain [5,10,19,20].
Figure 2.QFISH with chromosome-enumeration DNA probe for chromosome 1 on nuclei of neural cells from the developing human brain. Nucleus 1: 2060, nucleus 2: 2180, nucleus 3: 4490 (relative intensities in pixels). Since nucleus 3 exhibit doubled intensity as to nuclei 1 and 2, left nucleus exhibit an association nut not chromosome loss.
Although previously mentioned approaches provide for effective detection of aneuploidy,their use does not allow to analyze whole interphase chromosomes in their integrity. The latter appears to be a major problem for all the studies related to interphase cytogenetics. To overcome this difficulty,an approach based on multicolor chromosome banding (for review see [15] and research articles of Dr. Liehr and associates cited thereof) for studying interphase chromosomes (interphase chromosomespecific multicolor banding or ICS-MCB) was elaborated [7,20]. ICS-MCB provides for studying chromosomes at all stages of cell cycle,at singlecell level and at molecular resolutions. To date,this is almost unique possibility for visualization of chromosomes in cells,which are not cultured in order to produce metaphase chromosomes (for more details concerning ICS-MCB see [7,20]).
In context of studying brain cells,this approach was found highly efficient and provided for assessment of natural chromosome number variation in the normal human brain,evaluation of aneuploidy in the developing human brain,and identification of aneuploidy as a pathogenic mechanism in human brain diseases (schizophrenia) [5,7,10,12,20].
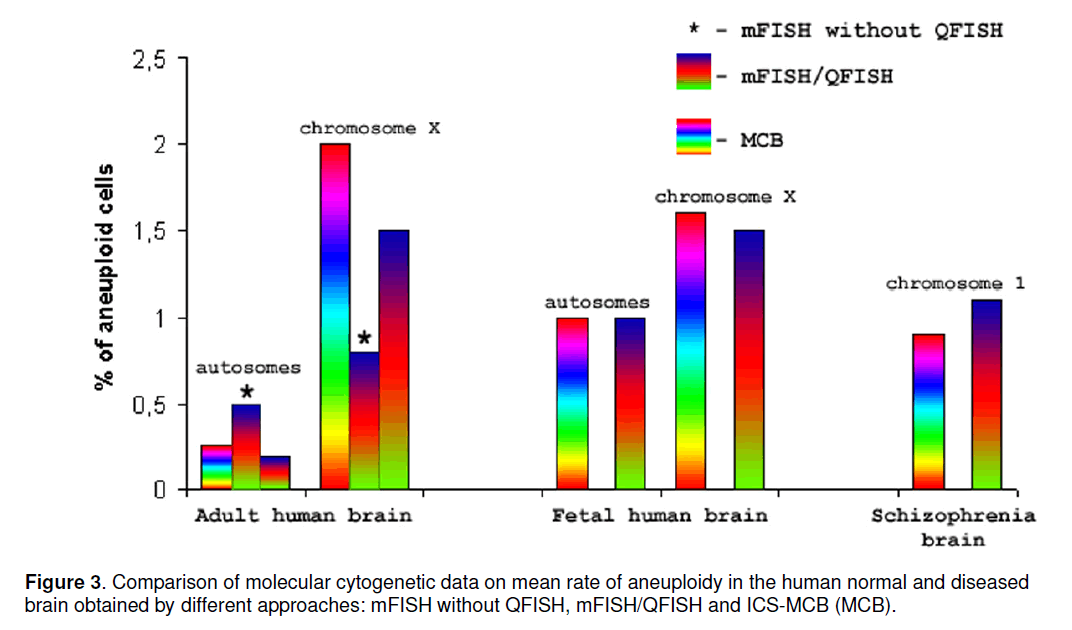
Since all aforementioned approaches are applicable for detection of aneuploidy in the human brain,it seems to be interesting to compare the data obtained. Figure 3 shows comparison of aneuploidy frequency (percentage of aneuploid cells) obtained by these approaches. The graph does not consider the cases of chromosome-specific aneuploidy increase that is observed in cases of schizophrenia (4 cases of aneuploidy for chromosomes 1,18 and X) [8,10] and is exceedingly common among fetal brain samples (up to 33%) [5]. One can see that mFISH/QFISH- and MCB-based approaches has comparable detection rate,whereas mFISH without QFISH causes autosomal aneuploidy overestimation and the X-chromosome aneuploidy underestimation. The latter is probably due to impossibility of differentiation between aneuploidy and peculiarities of chromosome organization in interphase nuclei of the brain. Nevertheless,ICS-MCB should be considered as more reliable approach inasmuch as having a look at the whole interphase chromosomes provides for identification of partial aneuploidy (i.e. loss/gain of a part of a chromosome) or additional rearranged chromosome. Although these types of chromosomal rearrangements were not observed in brain samples,as yet,studied,related chromosome imbalances could be present in cases of neuropsychiatric or neurodegenerative disease and in brain tumors [3]. Probes for ICS-MCB are less available as to commercial ones for mFISH/QFISH. Therefore,the concordance of data obtained on the developing,normal and schizophrenia brain by these techniques is rather encouraging in context of further molecular neurocytogenetic studies. However,to monitor chromosome instability in the human brain,it is to apply ICS-MCB.
4. Technical problems of FISH-based technique applications
Regardless molecular cytogenetic developments directly elaborated for brain cell studies,some problems remain. One of them is related to preparation of cell suspensions for further molecular cytogenetic analysis. Current approaches are elaborated for studying aneuploidy in interphase nuclei and use well-known interphase cytogenetic protocols [21]. However,these ones are poorly applicable for combination of FISH and other cytochemical or histochemical techniques targeted at labelling different types of brain cells (i.e. neuronal and glial cells or NSCs). Therefore,this point should be worked out through development of such composite approaches. Another problem frequently faced during mFISH analysis is related to extreme variability of DNA size within targeted chromosome regions. In rare cases,FISH signal lack is observed in interphase nuclei suggesting chromosome loss,whereas analysis of metaphase chromosomes does not reveal aneuploidy [22]. Here,the application of ICS-MCB is the only way to perform the analysis. Additional technical drawback of interphase FISH is referred to replication of chromosomal loci,that appears as two signals with identical intensities [23]. ICS-MCB provides for solution of this problem and QFISH can also help in overcoming this difficulty (for further review see [3,12]).
5. Alternative molecular genetic or cytogenetic techniques for studying intercellular genomic variations
FISH-based techniques remain the most popular for studying interphase nuclei of human somatic tissues [3,12,13]. However,there are alternatives that are molecular cytogenetic and molecular genetic techniques. The last decade offered a possibility of molecular cytogenetic analysis of the human genome at exceedingly high resolution. This set of techniques uses combination of comparative genomic hybridization (CGH) and microarray technologies and is termed array CGH (for review see [24]). Although the technique is primarily developed for studying genome using the total DNA isolated from a pool of cells,single-cell array CGH can be used for studying intercellular genomic variations [25]. Furthermore,recent developments in molecular genetics allowed to perform sequencing of DNA isolated from a cell [26]. The approach is primarily targeted at analysis of nucleotide sequence variation but can be still applied for detection of chromosomal imbalances. An approach,that analyses single nucleotide polymorphisms (SNP) by an array technology,was also found applicable for analyzing chromosomal alterations at single-cell level [27]. These techniques provides alternative to FISH-based ones. However,their application met a number of problems,which are not completely solved. The main one is related to necessity of whole-genome amplification that can be non-random in different genome regions causing high overestimation of genomic imbalances (for further review see [12,13]).
6. Combination of molecular cytogenetic techniques with other approaches for studying NSCs
Almost all studies described were essentially performed on the total cell fractions directly isolated from the developing or adult human brain. However,to perform a successful molecular cytogenetic study of NSCs,one should combine molecular cytogenetic techniques with approaches allowing to differentiate NSCs and other brain cells. Applied neurosciences possess numerous approaches that could be applied for detection of NSCs (for review see [1,2]),but there has no been a report describing the simultaneous. A study of aneuploidy in the adult human brain has applied immunofluorescence cell sorting sequentially followed by application of twocolour- FISH [6]. However,it is to note that aneuploidy detection in the normal adult brain is a scoring of rare events. Therefore,the frequency of aneuploidy in the normal human brain can be less than resolution of cell sorting (i.e. non-isolated cells achieve the rate of above 5%,whereas chromosome-specific aneuploidy in the normal adult brain rarely exceeds 1%) [1-14]. Thus,to obtain better resolution,one should use simultaneously molecular cytogenetic techniques with cytochemical and histochemical approaches for labelling NSCs. Here,two main approaches appear to be available:
(i) FISH or FISH-based techniques with either immunofluorescence or non-fluorescent techniques for marking NSCs; (ii) immunofluorescence detection of NSCs followed by interphase nucleus isolation and an alternative technique (such as CGH). To elaborate these approaches,one should consider several points: (i) use of fluorescence for cyto/histochemical detection of NSCs diminishes the resolution of mFISH approaches (especially ICS-MCB) because of reduction of at least one fluorescence channel; (ii) cytochemical techniques without fluorescence (light microscopy) negatively affect FISH results through diminishing fluorescence intensity; (iii) differences between cell processing for FISH or other molecular cytogenetic techniques and cyto/histochemical labelling of NSCs. Hopefully,forthcoming studies will consider this points and develop an integrated technique for detection of aneuploidy in labelled NSCs.
7. Conclusion
NSC biology has come to encompass different areas of biomedical research. The importance of studying NCS properties and behaviour has been repeatedly underlined [1,2]. Additional implications of NSC research are referred to their use as substrate for therapy of human devastative neurological diseases,which are exceedingly heavy burden for patients and their relatives [28]. Moreover,a number of studies evidences for NSC contribution to brain tumorigenesis [29]. This suggests aneuploidy detection in NSCs to have importance for the delineation of brain tumour origin. The latter becomes even more interesting in the light of recent findings demonstrating that aneuploidy can act both oncogenically and as a tumor suppressor,which is common for tumors of different tissues [30]. Furthermore,related phenomena (progressive aneuploidization,aneuploid cell persistence) are assumed to be involved in human neurogenesis during prenatal development [5]. Consequently,studying aneuploidy in NSCs has multilateral relevance. State-of-the-art molecular cytogenetic techniques provide for high-resolution aneuploidy detection in NSCs. However,to increase the sensitivity of molecular cytogenetic NSC research,new enhancements of the established techniques seem to be required. The present communication focuses on the main challenges related to aneuploidy detection and depict advantages and disadvantages of molecular cytogenetic techniques in relation to investigations of the human brain in health and disease. Hopefully,these data can serve a basis for designing future approaches towards chromosome studying in NSCs and appropriate application of molecular cytogenetic techniques that will lead to new discoveries in the NSC research.
Acknowledgements
We are indebted to Dr. Ilia Soloviev for contribution to development of molecular cytogenetic techniques in our laboratories. Authors are supported by Philip Morris USA Inc. and a grant LI820/18-1 from DFG (to IY Iourov).
References
- Gage F.H. (2000) Mammalian neural stem cells. Science,289(5457): 1433-1438.
- Ming G.L.,Song H. (2005) Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci,28: 223-250.
- Iourov I.Y.,Vorsanova S.G.,Yurov Y.B. (2006) Chromosomal variations in mammalian neuronal cells: known facts and attractive hypotheses. Int Rev Cytol,249: 143-191.
- Yurov Y.B.,Iourov I.Y.,Monakhov V.V.,Soloviev I.V.,Vostrikov V.M.,Vorsanova S.G. (2005) The variation of aneuploidy frequency in the developing and adult human brain revealed by an interphase FISH study. J Histochem Cytochem,53(3): 385-390.
- Yurov Y.B.,Iourov I.Y.,Vorsanova S.G.,Liehr T.,Kolotii A.D.,Kutsev S.I.,Pellestor F.,Beresheva A.K.,Demidova I.A.,Kravets V.S.,Monakhov V.V.,Soloviev I.V. (2007) Aneuploidy and confined chromosomal mosaicism in the developing human brain. PLoS ONE,2(6): e558.
- Rehen S.K.,Yung Y.C.,McCreight M.P.,Kaushal D.,Yang A.H.,Almeida B.S.V.,Kingsbury M.A.,Cabral K.M.S.,McConnell M.J.,Anliker B.,Fontanoz M.,Chun J. (2005) Constitutional aneuploidy in the normal human brain. J Neurosci,25(9): 2176-2180.
- Iourov I.Y.,Liehr T.,Vorsanova S.G.,Kolotii A.D.,Yurov Y.B. (2006) Visualization of interphase chromosomes in postmitotic cells of the human brain by multicolour banding (MCB). Chromosome Res,14(3): 223-229.
- Yurov Y.B.,Vostrikov V.M.,Vorsanova S.G.,Monachov V.V.,Iourov I.Y. (2001) Multicolor fluorescent in situ hybridization on post-mortem brain in schizophrenia as an approach for identification of low-level chromosomal aneuploidy in neuropschychiatric diseases. Brain Dev,23(S1); 186-190.
- Yurov Y.B.,Vorsanova S.G.,Iourov I.Y.,Demidova I.A.,Beresheva A.K.,Kravetz V.S.,Monakhov V.V.,Kolotii A.D.,Voinova-Ulas V.Y.,Gorbachevskaya N.L. (2007) Unexplained autism is frequently associated with low-level mosaic aneuploidy. J Med Genet,44(8): 521-525.
- Yurov Y.B.,Iourov I.Y.,Vorsanova S.G.,Demidova I.A.,Kravetz V.S.,Beresheva A.K.,Kolotii A.D.,Monakchov V.V.,Uranova N.A.,Vostrikov V.M.,Soloviev I.V.,Liehr T. (2008) The schizophrenia brain exhibits low-level aneuploidy involving chromosome 1. Schizophr Res,98(1-3): 137-147.
- Yurov Y.B.,Vorsanova S.G.,Vostrikov V.M.,Monakhov V.V.,Soloviev I.V.,Iourov I.Y. (2006) In vitro cultivation of fetal brain cells induces aneuploidy: a caution for neural stem cell therapy? Int J Neuroprotec Neuroregenerat,2(3): 209-211.
- Iourov I.Y.,Vorsanova S.G.,Yurov Y.B. (2006) Intercellular genomic (chromosomal) variations resulting in somatic mosaicism: mechanisms and consequences. Curr Genomics,7(7): 435-446.
- Kingsbury M.A.,Yung Y.C.,Peterson S.E.,Westra J.W.,Chun J. (2006) Aneuploidy in the normal and diseased brain. Cell Mol Life Sci,63(22): 2626-2641.
- Muotri A.R,Gage F.H. (2006) Generation of neuronal variability and complexity. Nature,441(7097): 903-910.
- Liehr T.,Starke H.,Weise A.,Lehrer H.,Claussen U. (2004). Multicolor FISH probe sets and their applications. Histol Histopathol,19(1): 229-237.
- Iourov IY.,Vorsanova S.G.,Yurov Y.B. (2008) Recent patents on molecular cytogenetics. Recent Patents DNA Gene Sequences,2(1): 6-15.
- Yurov Y.B.,Soloviev I.V.,Vorsanova S.G.,Marcais B.,Roizes G.,Lewis R. (1996) High resolution fluorescence in situ hybridization using cyanine and fluorescein dyes: ultra-rapid chromosome detection by directly fluorescently labeled alphoid DNA probes. Hum Genet,97(3): 390-398.
- Vorsanova S.G.,Kolotii A.D.,Iourov I.Y.,Monakhov V.V.,Kirillova E.A.,Soloviev I.V.,Yurov Y.B. (2005) Evidence for high frequency of chromosomal mosaicism in spontaneous abortions revealed by interphase FISH analysis. J Histochem Cytochem,53(3): 375-380.
- Iourov I.Y.,Soloviev I.V.,Vorsanova S.G.,Monakhov V.V.,Yurov Y.B. (2005) An approach for quantitative assessment of fluorescence in situ hybridization (FISH) signals for applied human molecular cytogenetics. J Histochem Cytochem,53(3): 401-408.
- Iourov I.Y.,Liehr T.,Vorsanova S.G.,Yurov Y.B. (2007) Interphase chromosome-specific multicolor banding (ICS-MCB): a new tool for analysis of interphase chromosomes in their integrity. Biomol Eng,24(4): 415-417.
- Iourov I.Y.,Vorsanova S.G.,Pellestor F.,Yurov Y.B. (2006) Brain tissue preparations for chromosomal PRINS labeling. Methods Mol Biol,334: 123-132.
- Vorsanova S.G.,Iourov I.Y.,Beresheva A.K.,Demidova I.A.,Monakhov V.V.,Kravets V.S.,Bartseva O.B.,Goyko E.A.,Soloviev I.V.,Yurov Y.B. (2005) Non-disjunction of chromosome 21,alphoid DNA variation,and sociogenetic features of Down syndrome. Tsitol Genet,39(6): 30-36.
- Soloviev I.V.,Yurov Y.B.,Vorsanova S.G.,Fayet F.,Roizes G.,Malet P. (1995) Prenatal diagnosis of trisomy 21 using interphase fluorescence in situ hybridization of postreplicated cells with site-specific cosmid and cosmid contig probes. Prenat Diagn,15(3): 237–248.
- Bejjani B.A.,Shaffer L.G. Application of array-based comparative genomic hybridization to clinical diagnosis. J Mol Diagn,8(5): 528-533.
- Le Caignec C.,Spits C.,Sermon K.,De Rucke M.,Thienpont B.,Debrock S.,Staessen C.,Moreau Y.,Fryns J.P.,Van Streirteghem A.,Liesbaers I.,Vermeesch J.R. (2006) Single-cell chromosomal imbalances detection by array CGH. Nucleic Acids Res,34(9): e68.
- Zhang K.,Martiny A.C.,Reppas N.B.,Barry K.W.,Malek J.,Chishholm S.W.,Church G.M. (2006) Sequencing genomes from single cells by polymerase cloning. Nat Biotech,24(6): 680-686.
- Iwamoto K.,Bundo M.,Ueda J.,Nakano Y.,Ukai W.,Hashimoto E.,Saito T.,Kato T. (2007) Detection of chromosomal structural alterations in single cells by SNP arrays: a systematic survey of amplification bias and optimized workflow. PLoS ONE,2(12): e1306.
- Lindvall O.,Kokaia Z. (2006) Stem cells for the treatment of neurological disorders. Nature,441(7097): 1094-1096.
- Vescovi A.L.,Galli R.,Reynolds B.A. (2006) Brain tumor stem cells. Nat Rev Cancer,6(6): 425-436.
- Weaver B.A.,Silk A.D.,Montagna C.,Verdier- Pinard P.,Cleveland D.W. (2007) Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell,11(1): 25-36.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences