Biocomputational Analysis of Filaggrin Sequence Repeats for Rheumatoid Arthritis
Sivakumar Prasanth Kumar, Saumya K. Patel, Yogesh T. Jasrai,, Himanshu A. Pandya, Pappu Srinivasan
1Department of Bioinformatics, Applied Botany Centre (ABC), University School of Sciences, Gujarat University, Ahmedabad - 380 009, India
2Department of Bioinformatics, Science Block, Alagappa University, Karaikudi - 630 004, India.
- Corresponding Author:
- Tel: +91 (0)79 26302578
Fax: +91 (0)79 26306194
E-mail: yjasrai@yahoo.com
Abstract
Anti-citrullinated antibodies are autoantibodies detected in the blood of rheumatoid arthritis (RA) patients in the early stage. These autoantibodies recognize an epitope of citrullinated peptides found on the surface of filaggrin protein and are cross-reactive in immune response. Since the expression of filaggrin protein is associated with autoantibodies secretion, it may act as a potential serological marker for the detection of RA in early stage. In the present study, the contribution of filaggrin sequence repeats towards antigenicity was investigated using bioinformatic approaches. The electrostatic potential of citrullinated filaggrin repeats and its antigenicity was found to be the prominent factors for invoking such an immune response.
Keywords
Anti-citrullinated antibodies, epitope, filaggrin sequence repeats, rheumatoid arthritis, electrostatic potential, antigenicity.
1. Introduction
Anti-citrullinated protein antibodies (ACPA) or anti-cyclic citrullinated peptide (anti-CCP) antibodies are autoantibodies directed against citrullinated proteins that are frequently detected in the blood of rheumatoid arthritis (RA) patients. The main epitope for these antibodies is the citrullinated peptides found on the surface of filaggrin protein [1]. The presence of autoantibodies against citrullinated proteins was first described in the mid-seventies when the biochemical basis of antibody reactivity against keratin and filaggrin was investigated [2]. According to American College of Rheumatology, approximately 95% of patients with a positive CCP test will have more chances of developing RA in the future. Hence, the detection of cross-reactivity between autoantibodies and filaggrin will act as a potential serological marker in the early detection of this disorder [3].
Filaggrin, the main protein of the keratohyalin granules, is produced during the last stages of mammalian epithelial cells terminal differentiation. In this stage, the protein is dephosphorylated and 20% of the arginine residues are converted into citrulline by the peptidylarginine deiminase enzyme present in the granules [4,5]. In the present study, the filaggrin sequence repeats was studied using sequence and structure-based approaches of bioinformatics to understand the antigenicity invoked by its expression.
2. Materials and methods
2.1 Sequence analysis of filaggrin repeats
The protein sequence of filaggrin (P20930) was retrieved from UniProtKB [6] and it was comprised of 23 sequence repeats of unequal length. The sequence repeats were then aligned using ClustalW algorithm accessible at EBI server [7] and insertions and deletions (INDELs) was studied with the help of sequence logo, created using Weblogo server [8]. A randomly chosen filaggrin sequence repeat was analyzed using SignalP 3.0 server [9] to identify whether the sequence has potential to act as signaling peptide. The prediction was executed using a model trained in artificial neural network (ANN) and hidden Markov model (HMM) with the search space limited to eukaryotes as organism type and with no N-terminal truncation. To study the antigenicity of filaggrin protein as whole and its repeats, Kolaskar and Tongaonkar antigenicity scale implemented in Immune Epitope Database and Analysis Resource (IEDB) [10] was utilized. This scale was based on semi-empirical method which makes use of physicochemical properties of amino acid residues and their frequencies of occurrence in experimentally known segmental epitopes to predict antigenic determinants on proteins [11]. Filaggrin complete sequence and its repeats were separately given as input.
2.2 Molecular modeling of filaggrin sequence repeats
An attempt was made to construct homology model using Swiss-Modeller [12]. But, no suitable template was identified with the default settings of InterproScan, Gapped Blast, and HH Template library search using the ‘Template Identification’ module. Hence, fold modeling was performed using PHYRE server [13]. The following were the steps in the procedure for fold modeling with a randomly selected filaggrin sequence repeat as input.
Secondary structure information was obtained based on a consensus generated by PsiPred, Jnet and SSpro programs. To find whether the queried sequence belongs to ordered (regular) structure or a component of disordered topology, it was examined via Disopred interface. Finally, PROSITE [14] was scanned to detect remote homologies for the query sequence and was modeled with the folds derived from Structural Classification of Proteins (SCOP) database. After assigning the Cartesian coordinates, the loops were modeled.
2.3 Molecular Citrullination of filaggrin repeats
Citrullination involves the terminal modifications of NH2 to =O group in arginine residues. Using YASARA View [15], NH2 group was initially deleted and terminal oxygen was added with the ‘Build’ utility. Bond orders were then corrected using ‘Adjust bond order’ utility. The modified (citrullinated) protein was saved in .pdb format and energy minimized using GRAMMX force field implemented in spdbv [12].
2.4 Generation of Electrostatic potential map
The electrostatic potential maps were generated for both the non- and citrullinated protein structure using PBEQ-Solver [16] which was based on Poisson-Boltzmann method. The electrostatic potential surface was created using Coulomb electrostatic method implemented in spdbv [12]. The generated maps were then analyzed using standard graphic visualizer.
3. Results and Discussion
3.1 Sequence analysis of filaggrin repeats
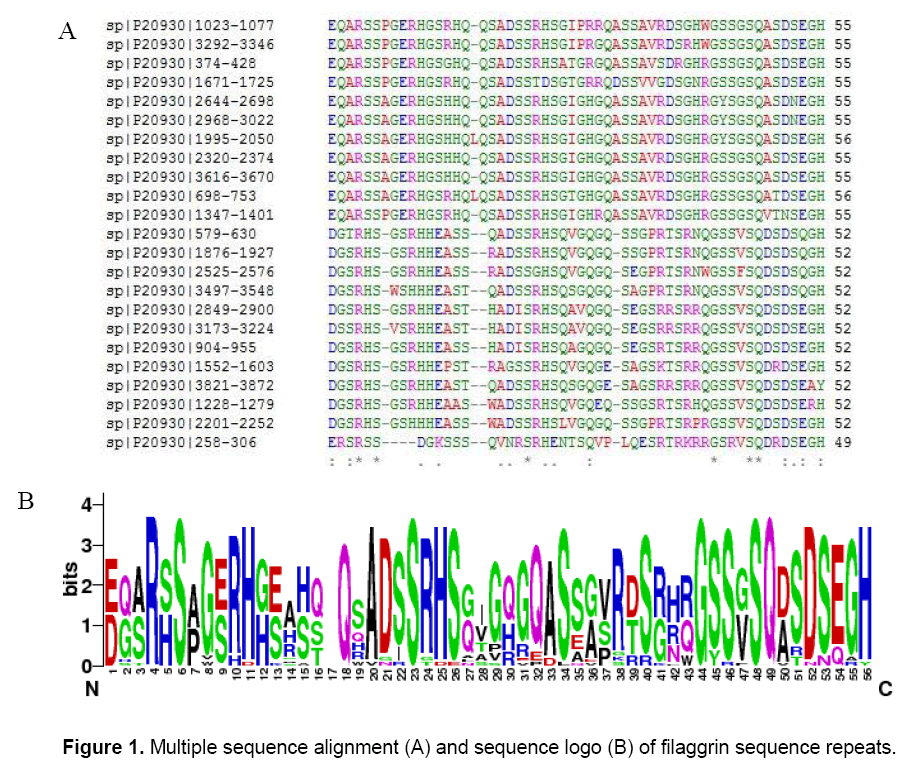
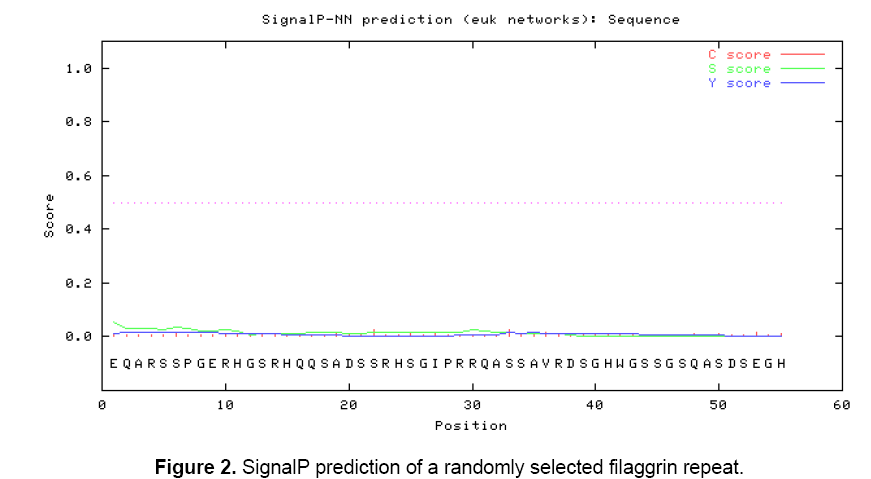
The objective is to determine whether the filaggrin sequence repeats contains antigenic properties or not and it was investigated through sequence and structure based approaches. In order to identify whether the repeats were identical or similar and the generated multiple sequence alignment (MSA) showed that there were many conserved and semi-conserved residues found in the repeats (Figure 1 A). The generated sequence logo made a clear picture to understand the conservative residues found in the columns: 4, 6, 8, 10, 11, 18, 23, 34, 40, 44, 45, 46, 48, 49, 52, 55 and 56. Significant INDEL was found in the position of 17 and 30. The higher text size in terms of bits depicted the overall conservativeness of filaggrin repeats (Figure 1 B). A randomly selected filaggrin repeat was scanned for potential signaling peptide to study whether it encompasses the function related to signaling pathways or not. SignalP 3.0 server predicted no signal peptide with the varied cut offs ranging from 0.32 to 0.43. The cleavage site score (C score), the signalling peptide score (S score) and the Y score (combination of C- and S score) were found to be linear representing no signaling peptides were found with the defined cut offs (Figure 2).
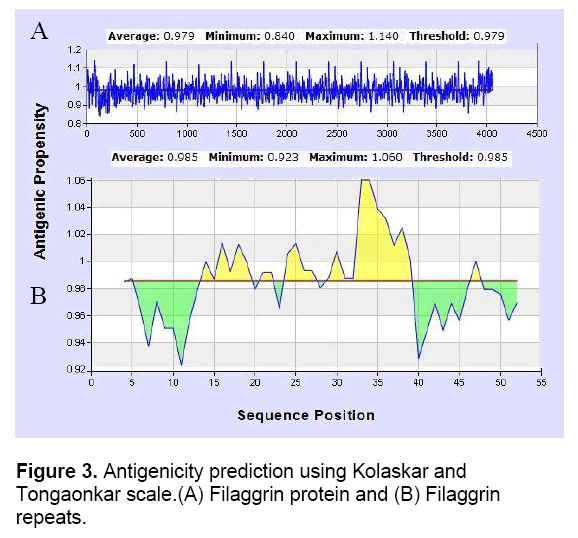
Thus, it was predicted that filaggrin repeat is a non-secretory protein. Regions of antigenicity in the filaggrin protein were analyzed using Kolaskar and Tongaonkar antigenicity scale with the threshold of 0.979. The antigenic propensity was observed with a minimum of 0.840 and maximum of 1.140 which indicated there are many potential epitopes present in the filaggrin protein (Table 1). A randomly selected filaggrin repeat was also specified as input to understand the contribution of antigenicity. The minimum and maximum value of 0.923 and 1.060 with the threshold setting of 0.985 showed that it act as a potential epitope while the threshold, minimum and maximum value were very close to each other.
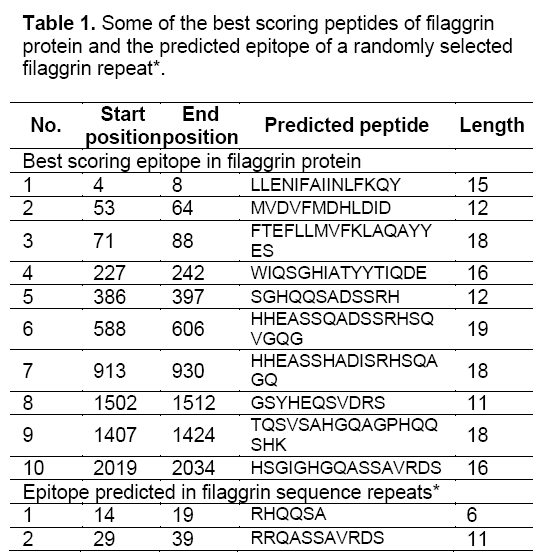
It was also observed that the signal peptide and signal anchoring probabilities were equal to zero which significantly increased the prediction accuracy. The peptides ‘RHQQSA’ and RRQASSAVRDS’ in the positions of 14-19 and 29-39 were predicted as epitopes (Figure 3). Arginine residues present in the filaggrin protein were converted to citrulline through structure modification. This event makes the protein act as citrullinated peptide (antigen) to mimic a condition in which the autoantibodies were secreted in RA disease. The predicted epitopes of filaggrin repeats contributed atleast one arginine residue which can potentially induce the filaggrin protein to act antigenic.
3.2 Molecular modeling of filaggrin sequence repeats
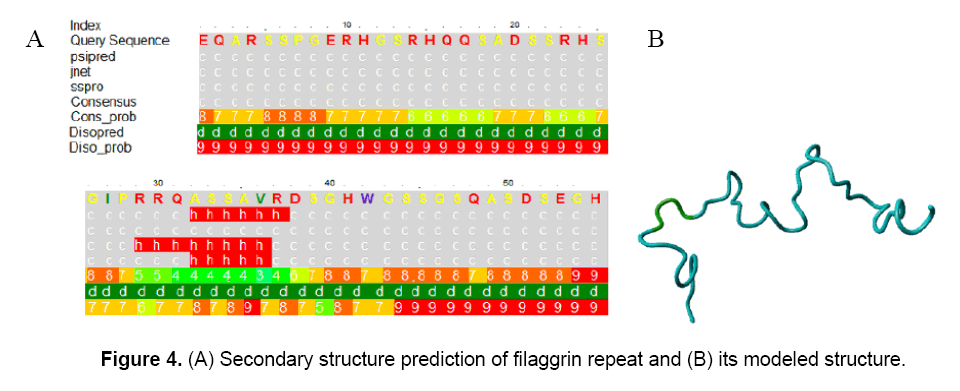
The secondary structure prediction of filaggrin repeat showed that it was composed of coiled-coil regions with a small stretch of alpha helix in the position of 32-36, respectively (Figure 4). The coiled-coil predicted region had highest probabilities of confidence which ranged from 7, 8 and 9.
However, the alpha-helix prediction was observed with a low consensus probability of 3 and 4. Thus, the structure was considered as a complete coiled-coil topology. The disorderness prediction of filaggrin repeat revealed that this topology is a highly disordered structure with disorder probability varied from 5 to 9 (Figure 4 A). Even PROSITE implemented in PHYRE could not able to detect distant homologs. The secondary structure based alignments were proved to be plausible. The filaggrin repeat was modeled with the fold derived from 2F3J chain A (SCOP code: c2f3ja) Structure of ref2-i mrna export factor 2. It was noticed that there was no sequence conservation observed in the cut offs expressed in percentage: 30%, 40%, 50% and 60% and predicted no evolutionary trace. The coiled-coil secondary structure alignment generated a model after loop modeling. After structure modification in arginine residues, the citrullinated protein was energy minimized to -1174.502 kJ/mol using GRAMMX force field (Figure 4 B).
3.3 Electrostatic potential maps
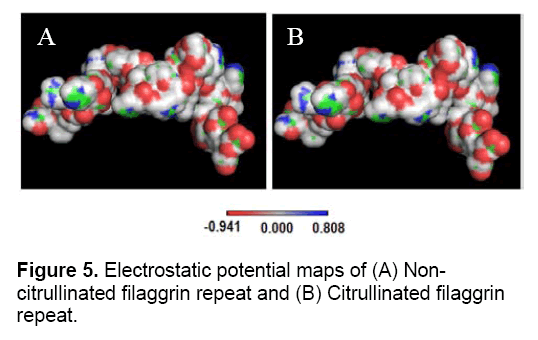
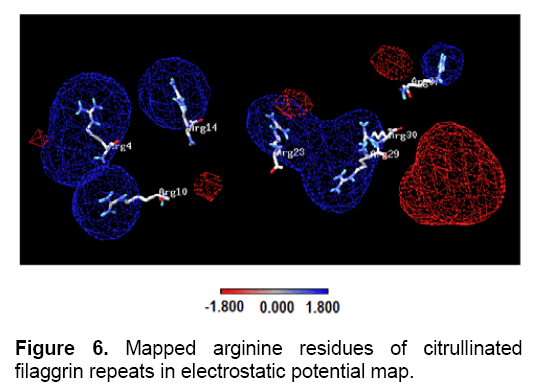
The antigen-antibody interaction is primarily facilitated through electrostatic interaction. Here, it was investigated how the electrostatic potential of filaggrin repeat invoke to act as an antigen. Electrostatic potential map was generated using PBEQ-Solver for both the filaggrin and citrullinated filaggrin repeats (Figure 5). The electrostatic potential surface was created using Coulomb electrostatic method with dielectric constant equal to 80.000 and based on charged residues, implemented in spdbv. Since arginine is a positively charged residue, it was mapped to positive region (charge distribution: 1.800). It should also be noted that the positive charged clusters (1.800) were dominated both in size and distribution in terms of polarity in contrast to negative charged clusters (-1.800) (Figure 6).
However, PBEQ-Solver provided an electrostatic potential map using Poisson-Boltzmann method which revealed uniform distribution of positive (0.808) and negative clusters (-0.941) with patches of neutrality. It should also be considered that surfaces represented in white color indicated that there is an equal energy and charge landscape throughout the molecule. The electrostatic map generated for both the non- and citrullinated proteins showed no significant changes in surface area. But, the computed solvation energy of non-citrullinated and citrullinated filaggrin repeats was found to be -1418.82 and -1411.33 Kcal/mol. The solvation energy is dependent on the arbitrary charge distributions over the protein molecules with the clear definition of impeded solvent dielectric continuum. The energy difference of 7.49 kcal/mol (variations in solvation energy) was proved to be evident in changing the molecular surface properties and contributed more towards the interaction of other protein molecules, especially an antibody.
4. Conclusions
The citrullination of arginine residues present on the filaggrin protein surface makes it to act as antigenic for which the autoantibodies were secreted reported in RA disease. Epitope prediction provided more informative sites which fortunately had atleast one arginine residue in its peptides. Structure based electrostatic potential map yielded many important insights. The molecular surface properties in terms of solvation energy are differed when the chemical (post-translational) modification of arginine to citrulline takes place. Thus, the correlation of sequence based epitope prediction and structure based electrostatic potential showed the significance of antigenicity. Citrullinated residue is observed to be embedded in positive charge cluster and it will be one of the properties required for electrostatic interaction with antibody. Thus, the present study had shed more insights into plausible biochemical and molecular aspects of filaggrin antigenicity which should be validated in molecular biology experiments.
Acknowledgements
SPK is supported by INSPIRE Fellowship, Department of Science and Technology, Govt. of India. Prof. Linz-buoy George is kindly acknowledged for reading the manuscript and provided valuable suggestions.
References
- Detrick B., Hamilton R.G., Folds J.D. (2006) Manual of molecular and clinical laboratory immunology. American Society for Microbiology Press, Washington, 1037-1043.
- American Association for Clinical Chemistry (2011) Labs test online. AACC Website: https://labtestsonline.org/understanding/analytes/ccp/tab/test. Retrieved on 14-7-2011.
- USCN Life Science Inc. (2009) Anti-filaggrin antibody (AFA). USCN Life Science Website: https://www.uscnk.com/directory/Anti-filaggrin-antibody(AFA)-1138.htm. Retrieved on 12-7-2011.
- Schellekens G.A., de Jong B.A.W., van den Hoogen, et al. (1998) Citrulline is an essential constituent of antigenic determinants recognized by Rheumatoid Arthritis–specific autoantibodies. J. Clin. Invest., 101: 273–281.
- Neuhauser E.G., Durieux J.J., Arnaud M., et al. (1999) The epitopes targeted by the Rheumatoid Arthritis-associated antifilaggrin autoantibodies are posttranslationally generated on various sites of (pro) filaggrin by deimination of arginine residues. J. Immunol., 162: 585-594.
- Boutet E., Lieberherr D., Tognolli M., et al. (2007) UniProtKB/Swiss-Prot - The manually annotated section of the UniProt KnowledgeBase. Meth.in Mol. Biol., 406: 89-112.
- Larkin M.A., Blackshields G., Brown N.P., et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics, 23: 2947-2948.
- Crooks G.E., Hon G., Chandonia, J.M., et al. (2004) WebLogo: a sequence logo generator. Genome Res., 14:1188-1190.
- Emanuelsson O., Brunak S., von Heijne G., et al. (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protoc., 2:953- 971.
- Peters B., Sidney J., Bourne P., et al. (2005) The Immune Epitope Database and Analysis Resource (IEDB): From vision to blueprint. PLoS Biol., 3:379-381. [11] Kolaskar A.S., Tongaonkar P.C. (1990) A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett., 276: 172-174.
- Guex N., Peitsch M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18: 2714-2723.
- Kelley L.A., Sternberg M.J.E. (2009) Protein structure prediction on the web: a case study using the Phyre server. Nature Protoc., 4:363-371.
- Hulo N., Bairoch A., Bulliard V., et al. (2006) The PROSITE database. Nucleic Acids Res., 34: 227–230.
- Krieger E., Joo K., Lee J., et al. (2009) Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins, 77:114-122.
- Jo S., Vargyas M., Szedlar J.V., et al. (2008) PBEQ-Solver for online visualization of electrostatic potential of biomolecules. Nucleic Acids Res., 36: 270–275.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences