Antioxidant Activities of Fungal Endophytes Isolated from Strawberry Fragaria x ananassa Fruit
Roland M. Hipol, Liezel M. Magtoto, Sigrid Minette A. Tamang and Amor M. Damatac II
Roland M. Hipo1*,Liezel M. Magtoto,Sigrid Minette A. Tamang and Amor M. Damatac2
Department of Biology,College of Science,University of the Philippines Baguio,Baguio City,Philippines
Abstract
Fragaria x ananassa (strawberry) fruit is one of the most common products of Baguio City and La Trinidad, Benguet, Philippines. In this study, strawberry fruits were sampled for its fungal endophytes. These organisms live within the living tissues of plants without causing any immediate and apparent negative effects. The antioxidant activities of fungal endophytes isolated and identified from strawberry fruits were evaluated. Fungal isolates were identified using ITSrDNA. Seven species of endophytes were successfully identified. These species were Sporidiobolus sp., Rhodotorula sp., Pilidium concavum, Corynespora cassiicola, Neodeightonia subglobosa, Aspergillus awamori, and Aspergillus sp. Antioxidant activities of these microorganisms were evaluated using ABTS decolorization assay and DPPH radical scavenging assay. Results showed that two species had antioxidant activities comparable to ascorbic acid. These isolates were Aspergillus awamori DT11 and Corynespora cassiicola DT13. Possibly, these fungal endophytes contribute to the total antioxidant system of the plant. Also, these fungal endophytes may also be contributors to the nutritive value of the strawberry fruits.
Keywords
Fungal endophytes; ITSrDNA; ABTS decolorization assay; DPPH radical scavenging assay; Total phenolic content.
1. Introduction
Strawberries (Fragaria x ananassa L.) are one of the most commonly consumed berries worldwide. They contain a large amount of phenolic compounds which have antioxidant properties. The high variety and content of polyphenolic constituents of strawberries,mainly represented by flavonoid anthocyanins and non-flavonoid condensed tannins,have recently attracted growing attention [1].
Antioxidants are substances that can prevent or delay oxidative damage of lipids,proteins,and nucleic acids by free radicals such as reactive oxygen species (ROS). ROS may cause disruption of membrane fluidity,denaturation of proteins,and peroxidation of lipids which could lead to cell damage and eventually death [2]. Researches for naturally occurring antioxidants have become a growing interest for the prevention and treatment of human diseases.
Any organism that is found inside the living tissues of plants is called an endophyte. Specifically,fungal endophytes are those that colonize living internal tissues of plants without causing any immediate and apparent negative effects [3]. They are ubiquitous and have been found in aboveground tissues,and in roots. They have been associated in all lineages in plants including liverworts,mosses,seed-free vascular plants,conifers,and angiosperms [4]. Some may receive nutrition and protection from host plant while the host plant may benefit from enhanced competitive abilities and increased resistance to herbivory,disease,and various abiotic stresses [5]. It was found that traits for drought tolerance,leaf chemistry,heavy metal tolerance in soils,and susceptibility of vegetative reproduction are directly attributable to the presence of endophytes [4]. There is an estimated 300,000 plant species that exist on earth,which could host one or more fungal endophyte [6]. However,only a few of them have been described to date and a lot are still waiting to be discovered. Many studies have shown that fungal endophytes produce natural products,which are bioactive that can be exploited as antibiotics,anticancer,anti-inflammatory,immunosuppressant and many other uses [7]. The discovery of this hidden diversity of fungi in plants may provide leads toward this end.
In this study,fruits of commercially grown strawberries from La Trinidad,Benguet were collected and from these,fungal endophytes were isolated and consequently identified through molecular means. Their antioxidant activities were also investigated.
2. Materials and Methods
Isolation of fungal endophytes
Fungal endophytes were obtained from the fruits of F. ananassa (strawberries). The fruits were taken at the Strawberry farm,La Trinidad,Benguet. Forty fruits were picked randomly from 40 different plants. The fruits were visually checked to make sure that there is no evidence of any manifestation of disease or infestation of insects. All fruits were sampled for endophytes within 24 hours of collection and were processed at the research laboratory of the University of the Philippines Baguio.
Fungal endophytes from these fruits were isolated according to methods used in Arnold & Lutzoni [4] with some modifications. Healthy fruits were washed in a running water to remove epiphytic debris and were dried. They were surface-sterilized using sequential immersion in 95% ethanol (1 minute),NaOCl solution (2:1 v/v water; 5 minutes),and 96% ethanol (30 seconds),sterilized water three times separately,and were allowed to surface dry under sterile conditions. A flame-sterilized knife was used to cut 10 pieces of 5x3x2 mm of tissue. They were plated immediately to potato dextrose agar (PDA). Two replicates were prepared for every fruit. To eliminate bacterial contaminants,an antibiotic powder 0·05gL-1 chloramphenicol was added to the PDA. A 0.005 gL-1 rose bengal was also added to retard the growth of fast-growing endophytic fungi. Plates were sealed,incubated at a temperature of 30°C in an oven incubator for 7-14 days. Emergent hyphae were transferred onto new PDA slants.
The endophytic fungal isolates were identified by sequencing the internal transcribed region (ITS) of the 18S rDNA,using universal primers ITS-1 (5’TCC GTA GAA CCT GCG G-3’) for the forward primer and ITS-4 (TCC TCC GCT TAT TGA TAT GC’) for the reverse primer. For this process,0.05 g of each pure culture was transferred to 1.5 microtubes for crude DNA extraction using KAPA express extract kit following manufacturer’s instructions. Afterwhich,1μL of the DNA extract was added to a PCR mixture consisting of 25 μL of KAPA3G Plant PCR buffer,25μL of PCR grade water and 1μL each for the forward and reverse primers. The reaction cycle consisted of a predenaturation phase of 5mins at 95°C,followed by 35 cycles of 30s at 95°C,1min at 60°C and 1min at 72°C. Final extension phase of 6mins at 72°C was also done. The PCR products were run on 1% agarose gel to check the amplification of the desired length which is about 550 bp. The PCR products were sent to Macrogen Inc. of Korea for cleaning and subsequent sequencing.
The Basic Local Alignment Sequence Tool for nucleotides (BLASTn) search program [9] was used to look for nucleotide sequence homology for the 18s,ITS (1/4) and 5.8s regions for the fungal isolates for their identification. Isolates whose sequences had a similarity greater than 95% were considered to belong to the same species. This 5% difference used to define species boundaries appears to correlate well with differences among known endophytic species [4]. In instances where the similarity is below 95%,only the genus was accepted.
Phylogenetic Analysis
DNA sequences of the isolates together with downloaded fungal sequences from the Genbank were subjected to Multiple Sequence Alignment algorithm,subsequent sequence editing and phylogenetic analysis using the Molecular Evolutionary Genetics Analysis software version 5.1 (MEGA) [8]. The same software was also used to determine the most appropriate model for the Maximum Likelihood algorithm for the derivation of the phylogenetic affinities of the studied sequences. Maximum Likelihood analysis used 500 bootstrap replications to come up with the consensus phylogenetic tree.
Fermentation and metabolite extraction
Fermentation and extraction of crude antioxidants were conducted according to methods by Zeng et al. [2]. All species were cultured in potato dextrose broth (PDB) for 7 days at 30°C with gentle shaking at 100 rpm using a rotary shaking incubator. After incubation,the broth of individual strains was filtered using Whatman #1. The supernatant obtained was extracted three times with ethyl acetate (v/v,1:1). The ethyl acetate was evaporated using reciprocal water bath set at 50 °C to yield the crude extract. The mycelium filtered was soaked in methanol for two days and filtered with Whatman #1. The methanol was evaporated using water bath set at 40 °C to yield the extract. In this study,the composite of the crude extracts for each endophytic fungi were used to determine their corresponding in-vitro antioxidant activities.
2,2’-Azino-di(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) decolorization assay
The ABTS decolorization assay was performed according to methods of Re et al. [9] with some modifications. ABTS stock solution (7mM) was reacted with potassium persulfate (2.45 mM) to produce the ABTS radical cation (ABTS+). The solution was stored for 12-16 hours under dark room temperature to become stable for more than two days. ABTS+ solution was diluted with doubledistilled water to achieve a concentration of 0.70±0.02 at 734 nm using UVmini-1240 UV-VIS Spectrophotometer. Thirty μL of sample extract was added to 3 mL of diluted ABTS+ solution. The absorbance was read every minute up to six minutes see the effect of the extract on ABTS+ against time. Ascorbic acid was used as positive control. Ascorbic acid equivalent of sample extracts was also determined using the % decolorization of ABTS of the different concentrations of ascorbic acid (2 μM- 12 μM) as the reference data.
2,2’-DiphenyL-1-picrylhydrazyl (DPPH) radical scavenging assay
The DPPH radical scavenging assay was performed according to methods by Miliauskas et al. [10] with slight modifications. One mL of sample extract (105 mg/mL) was added to 2 mL DPPH (0.004 g/mL). The mixtures were incubated at room temperature under dark for 30 minutes. Then,the absorbance was read at 517 nm. The scavenging ability on DPPH radicals were calculated using the equation
Scavenging ability on DPPH radicals (%) = [(A1 - A2) / A1] x 100; where A1 is the absorbance of the control,which contain all reagents except the sample extract,and A2 is the absorbance of the sample extract. Ascorbic acid was also used as positive control.
Statistical Analysis
Antioxidant assays were performed in triplicate. Statistical analysis was carried out using SPSS version 18 using ANOVA (P<0.05).
3. Results
3.1 Identification and Phylogeny of Isolates
The genus and species of the isolates were determined by sequencing the PCR products of each fungal endophytes using the primers ITS1 and ITS4. The identified species were: Sporidiobolus sp.,Rhodotorula sp.,Pilidium concavum,Aspergillus niger,Aspergillus awamori,Corynespora cassiicola,Neodeightonia subglobosa,Aspergillus sp. (Table 1). Among the identified species,5 were ascomycetes while only two were basidiomycetes.
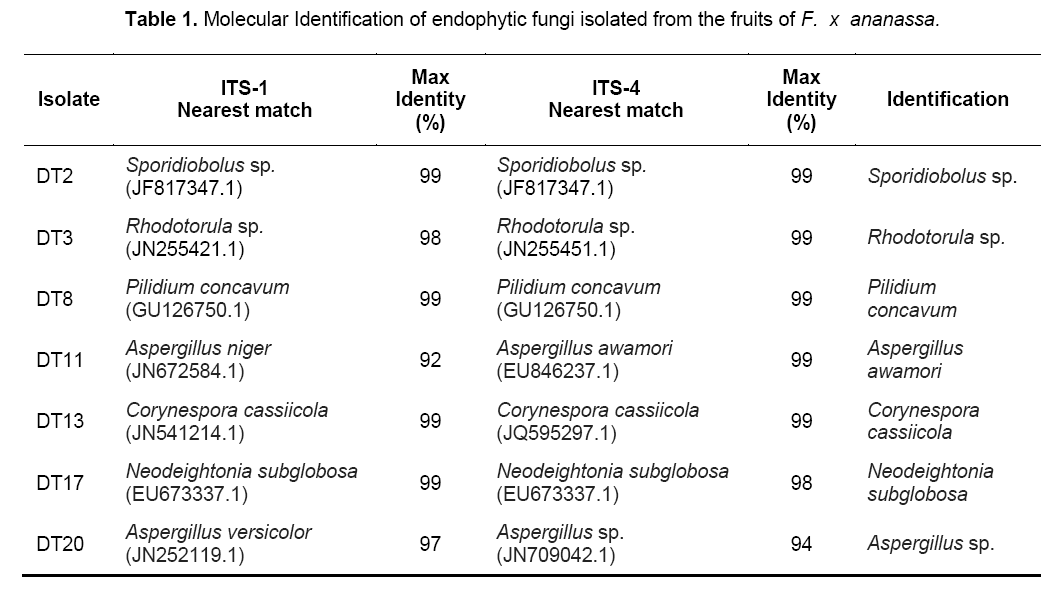
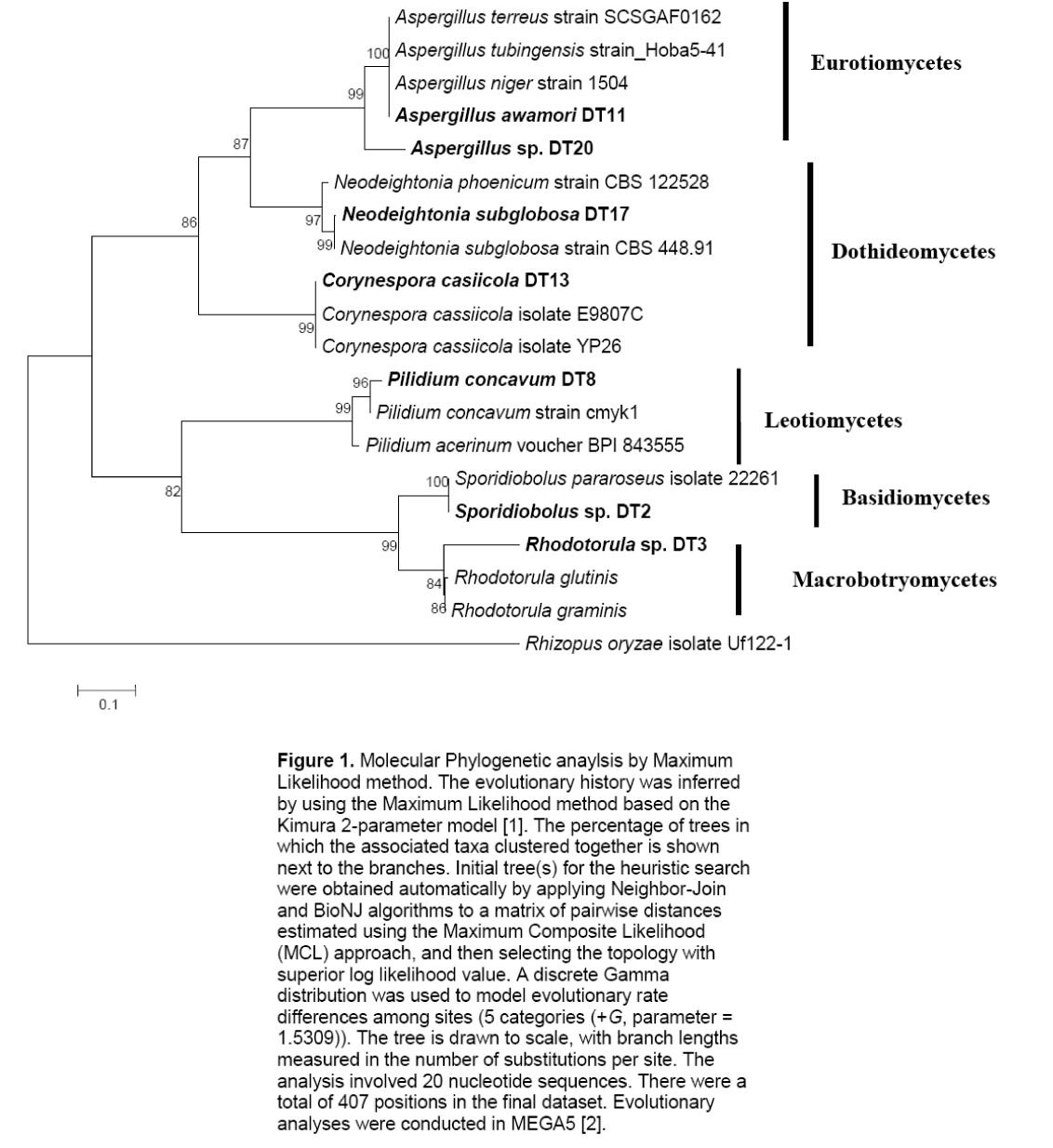
The two basidiomycetes were Sporidiobolus sp.,and Rhodotorula sp.; both of which are yeasts. The ITS sequences of the isolates were submitted to GenBank sequence database of the National Center for Biotechnology Information (NCBI) of the National Institutes of Health in the United States. Phylogenetic analyses showed five major subclades (Figure 1). The Ascomycete clade is represented by three classes namely Eurotiomycetes,Dothidiomycetes and Leotiomycetes. The Basidiomycete clade,clustering Sporidiobolus sp. and Rhodotorula sp. is also shown.
Figure 1: Molecular Phylogenetic anaylsis by Maximum Likelihood method. The evolutionary history was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model [1]. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 1.5309)). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 20 nucleotide sequences. There were a total of 407 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 [2].
3.2 Antioxidant Activity
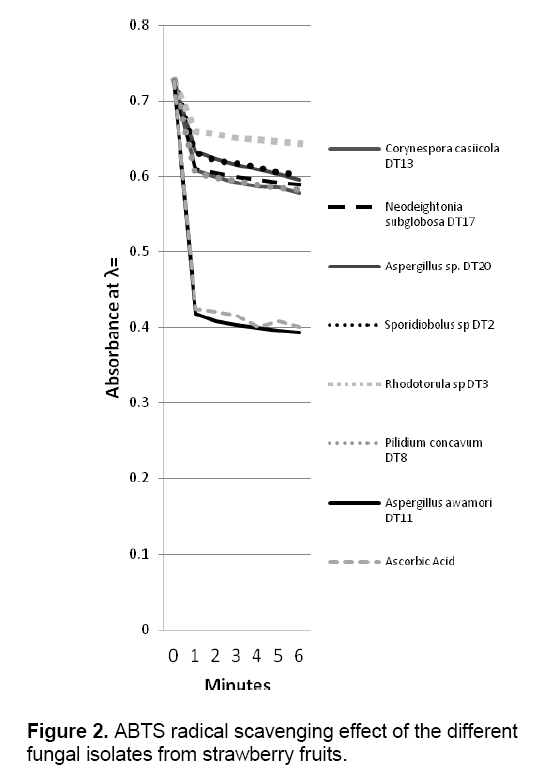
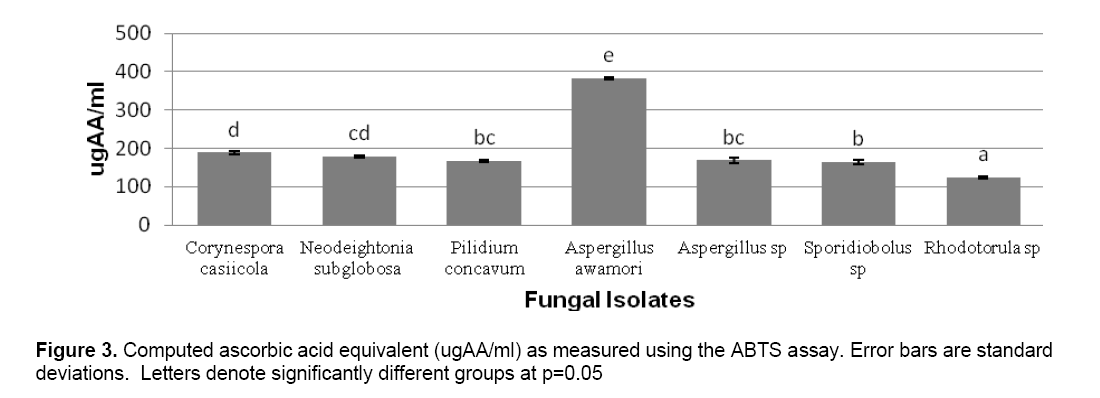
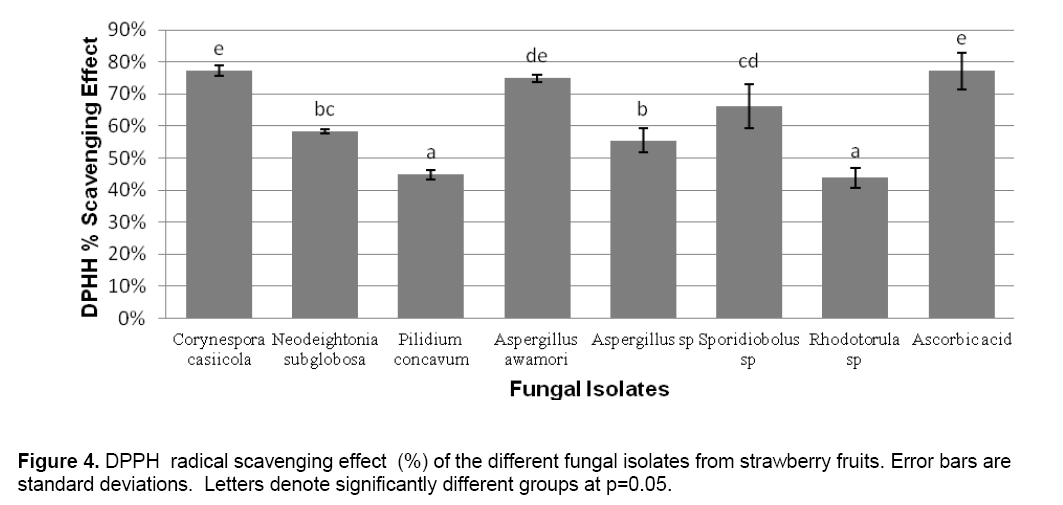
ABTS decolorization assay showed the effects of samples on the ABTS+ radical cation versus time (Figure 2 ). The results showed that the species that exhibited the greatest scavenging effects was Aspergillus awamori DT11 at 34.07% as compared to the positive control ascorbic acid which was 44.5%. Figure 3 shows the ascorbic acid antioxidant capacity of fungal endophytes against ABTS+ radical cation expressed in terms of equivalents. This was computed using the ascorbic acid assay generated standard curve (y = -31.144x + 23.827). Aspergillus awamori DT11 showed the highest ascorbic acid equivalent of 383.84 μg AA/ml of the extract. This value is 43% more than the average AA equivalents of all the other endophytes. On the other hand,the DPPH value showed two promising isolates: Corynespora casiicola DT13 and Aspergillus awamori DT11 (Figure 4 ). The % scavenging activity was measured to be 77.3%,74.15% respectively.
Figure 1. Molecular Phylogenetic anaylsis by Maximum Likelihood method. The evolutionary history was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model [1]. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach,and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G,parameter = 1.5309)). The tree is drawn to scale,with branch lengths measured in the number of substitutions per site. The analysis involved 20 nucleotide sequences. There were a total of 407 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 [2].
4. Discussion
Fungal endophytes are believed to be present in almost all plants [4]. To date,the most commonly isolated fungal endophytes are ascomycetes belonging to the Subphylum Pezizmycotina specifically to the subclades of Pezizomycetes,Leotiomycetes,Eurotiomycetes,Sordariomycetes,and Dothideomycetes [11,12]. The identified fungi in this research belong to the genera of Aspergillus,Pilidium,Corynespora and Neodeightonia; belonging to the Eurotiomycetes,Leotiomycetes,and Dothideomycetes,respectively. Basidiomycetous yeasts were also represented. The identified genera of Sporidiobolus and Rhodotorula both belong to subphylum Pucciniomycotina. The generated phylogenetic tree displays the affinitiesof the isolated fungal endophytes (Figure 1).
In the present study,the antioxidant activities of endophytic fungi were evaluated by two assays; ABTS and DPPH assays. Results show that all of the fungal endophyte isolates had antioxidant activities. The isolate of greatest interest,however,was Aspergillus awamori DT11. It showed high antioxidant activity in comparison to the other isolates.
Several other fungal representatives have been tested for antioxidant activity [13-16]. This activity is postulated to be the effect of the presence of several secondary metabolites including phenolic acids and their derivatives,flavonoids and phenolic terpenoids [16]. The isolated fungal endophytes from strawberry fruits may have these antioxidant compounds in abundant quantities,especially Aspergillus awamori DT11 resulting to noticeable ABTS/DPPH antioxidant activity. These antioxidant compounds produced by these endophytes may function to augment the antioxidant system of the host strawberry plant against possible biotic and abiotic stresses. Also,the endophytes are possibly contributing to the total nutritive value of the strawberry fruit. These results may also suggest that these endophytic fungi may be potential sources of natural antioxidants.
This is the first time that fungi endophytic to strawberry fruits are investigated for their antioxidant activities. However,the toxicity of the endophyte extracts should also be investigated,to check their safety as food additives [17]. It is also recommended that further research regarding the antioxidant constituents in the extracts of endophytic fungal isolates be pursued.
Acknowledgements
The authors would like to thank the University of the Philippines Baguio for the support in the conduct of this study.
References
- Seeram N.P.,Lee R.,Scheuller H.S.,& Heber D. (2006). Identification of phenolic compounds in strawberries by liquid chromatography electrospray ionization mass spectrometry. Food Chem.,97: 1-11.
- Zeng P.,Wu J.,Liao L.,Chen T.,Wu J.,& Wong K. (2011). In vitro antioxidant activities of endophytic fungi isolated from liverwort Scapania verrucosa. Genet Mol Res.,10(4): 3169-3179.
- Owen N.,& Hundley N. (2004). Endophytes-the chemical synthesizers inside plants. Sci. Prog.,87: 79-99.
- Arnold A.,& Lutzoni F. (2007). Diversity and Host Range of Foliar Fungal Endophytes: Are Tropical Leaves Biodiversity Hotspots? Ecology,88: 541-549.
- Saikkoken K.,Faeth S.,Helander M.,& Sullivan T. (1998). Fungal Endophytes: A Continuun of Interactions with Host Plants. Annu. Rev. Ecol. Evol. Syst.,29: 319-343.
- Strobel G.,& Daisy B. (2003). Bioprospecting for Microbial Endophytes and Their Natural Products. Microbiol. Mol. Biol. Rev. ,67: 491–502.
- Priti V.,Ramesha B.,Singh S.,Ravikanth G.,Ganeshaiah K.,Suryanarayanan T. and Shaanker R. (2009). Opinion: How promising are endophytic fungi as alternative sources of plant secondary metabolites? Curr. Sci.,97: 447-448.
- Tamura K,Peterson D.,Peterson N.,Stecher G.,Nei M.,and Kumar S. (2011). MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood,Evolutionary Distance,and Maximum Parsimony Methods. Mol. Bio. Evol.,28: 2731-2739.
- Re R.,Pellegrini N.,Proteggente A.,Pannala A.,Yang M.,& Rice-Evans C. (1999). Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med.,26: 1231-1237.
- Miliauskas G.,Venskutonis P.,& Van Beek T. (2004). Screening of radical scavenging activity of some medicinal and aromatic plant extracts. Food Chem.,85: 231-237.
- Lutzoni F.,Kauff F.,Cox,C.,McLaughlin D.,Celio G.,Dentinger B. (2004). Assembling the fungal tree of life: progress,classification,and evolution of subcellular traits. Am J Bot. ,91: 1446-1480.
- James T.,Kauff F.,Schoch C.,Matheny P.,Hofstetter V.,Cox C.,et al. (2006). Reconstucting the early evolution of the fungi using a six gene phylogeny. Nature,443: 818-822.
- Murthy N. K.,Pushpalatha,K. C.,& Joshi,C.G. (2011). Antioxidant activity and phytochemical analysis of endophytic fungi isolated from Lobelia nicotianifolia. J Chem Pharm Res.,3(5): 218-225.
- Ravindran C.,Naveenan T.,Varatharajan G.R.,Rajasabapathy R.,& Meena R.M. (2012). Antioxidants in mangrove plants and endophytic fungal associations. Bot. Mar.,55: 269–279.
- Cui Y.,Kim D.S.,& Park K.C. (2005). Antioxidant effect of Inonotus obliquus. J. Ethnopharmacol.,96: 79-85.
- Kumar S.,Dhankhar S.,& Yadav J.P. (2012). Antioxidant activity of fungal endophytes isolated from Salvadora oleoides Decne. Int. J Pharm. Pharm. Sci.,4(2):,380–385.
- Liu X.,Dong M.,Chen X.,Jiang M.,Lu X.,& Yan G. (2007). Antioxidant activity and phenolics of an endophytic Xylaria sp. from Ginkgo biloba. Food Chem.,105: 548–554.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences