A Mini Review on Some Aspects of the Biochemistry of the Micronutrient Molybdenum (VI), (Mo6+)
Yahia Z Hamada*, Alana Antoine
Published Date: 2018-04-30Yahia Z Hamada*, Alana Antoine
Division of Natural and Mathematical Sciences, LeMoyne-Owen College, 807 Walker Avenue, Memphis, TN 38126 USA
- Corresponding Author:
- Yahia Z Hamada
Division of Natural and Mathematical Sciences
LeMoyne-Owen College, 807 Walker Avenue, Memphis, TN 38126 USA
Tel: 1(901) 435-1392
E-mail: Yahia_hamada@loc.edu
Received date: March 20, 2018; Accepted date: April 11, 2018; Published date: April 30, 2018
Citation: Hamada YZ, Antoine A. A Mini Review on Some Aspects of the Biochemistry of the Micronutrient Molybdenum (VI), (Mo6+) Electronic J Biol, 14:2
Abstract
This mini-review touches on some of the most important aspects of the biochemistry of the second row transition metal, micronutrient, molybdenum (VI) (Mo6+). Molybdenum can exist in metal complexes in a variety of oxidation states ranging from the metallic oxidation state of 0 to the most oxidized form of +6. To date, it is believed that molybdenum is taken up by living cells as the molybdate anion [MoO4]2-. There are not many reviews in the literature that cover the current topic. There are a total of about 50 Mo- Containing Enzymes/Proteins. Alongside the detailed literature review, we are also presenting the reactions of aqueous Mo6+ with the organic ligand Malic Acid (MA). It appeared that, the reaction of Mo6+ with MA in aqueous solutions at 25°C in 0.1 M ionic strength (NaNO3) formed a reaction mixture that released a large number of hydrogen ions, or protons (H+); 17 H+ to be exact. This observation is not surprising for such complex behavior of such complex metal ion in aqueous solutions. This mini-review is a contribution to celebrate the 85th birthday of Professor Mostafa El-Sayed; at department of chemistry of the Georgia Institute of Technology, Atlanta, Georgia, USA.
Keywords
Aqueous solutions; Molybdenumcontaining- enzymes; Malic acid; Mo6+; Nitrogenase; Potentiometry.
Introduction
Among all micronutrients, Molybdenum possesses very unique characters. It is the only second row transition metal that has a tangible biological activity, it exists in a wide variety of oxidation states (ranging from 0 to +6) and it is a required co-factor for at least four dozen enzymes [1]. There are a limited number of reviews or mini-reviews that have appeared with the biology/biochemistry of molybdenum in mind [1- 3], particularly in aqueous solutions. However, the meticulous and thorough 75 page review by Hille et al. [2] is a great reference for the biochemistry of molybdenum of which they cited 536 other biomolybdenum- related research articles.
Herein, we have conducted detailed literature research to prepare for this min-review and found the following three facts: (1) Not many research articles dealt with the reaction of Mo6+ and aqueous solutions; (2) The chemistry of molybdenum is extremely complex; and (3) Billions of years ago, nature understood the uniqueness of molybdenum biochemistry that scientists only recently have recognized [1-20]. Figure 1 of the supplementary materials shows the details of this library search of the molybdenum research articles and review articles that were found within all American Chemical Society (ACS) Journals. It is noteworthy that there are a total of 44 journals within all ACS publication domains, which publishes thousands of research papers and reviews monthly.
There are at least four dozens known molybdenumcontaining- enzymes and or proteins (molybdoenzymes); Nitrogenase being the most well-known among all of them within the biology and chemistry audience. Herein, we are going to mention a dozen as example of these known molybdo-enzymes: (1) Nitrogenase, (2) Nitrate Reductase, (3) Xanthine Oxidase or Xanthine Dehydrogenase, (4) Pyrimidine Oxidase/Aldehyde Oxidase, (5) Trimethylamine Oxide Reductase, (6) Formate Dehydrogenase (7) Carbon Monoxide Oxoreductase/Carbon Monoxide Dehydrogenase, (8) Pyridoxal Oxidase, (9) Sulfite Oxidase, (10) Biotin Sulfoxide Reductase, (11) Dimethyl Sulfoxide Redutase, and (12) Tetrathionite Reductase. Table 1 catalogues all of these enzymes. Some of these Molybdenum-containingenzymes were isolated from bacteria (particularly cyanobacteria), fungi, yeast, plants, or mammals. For more details refer to references 1-3 and all 694 references mentioned therein. This current mini-review will focus on the discussion of the Molybdenum-containing-enzymes “Nitrogenase”.
| Serial No. | Enzyme/protein | Enzyme Commission # (EC) | Action/Function |
|---|---|---|---|
| 1 | Nitrogenase | 1.18.6.1 | Converts nitrogen to ammonia (N2 to NH3) |
| 2 | Nitrate Reductase | 1.7.99.4 | Converts nitrate to nitrite (NO3- to NO2-) |
| 3 | Xanthine Oxidase | 1.17.3.2 | Converts Xanthine to Uric acid |
| 4 | Pyrimidine Oxidase/Aldehyde Oxidase | 1.2.3.1 | Converts Aldehydes to Carboxylic acids |
| 5 | Trimethylamine Oxide Reductase | 1.6.6.9 | Converts Trimethylamine-N-Oxide to trimethylamine |
| 6 | Formate Dehydrogenase | 1.2.1.2 | Converts Formate to CO2 |
| 7 | Carbon Monoxide Oxoreductase/Carbon Monoxide Dehydrogenase | 1.2.99.2 | Converts CO to CO2 |
| 8 | Pyridoxal Oxidase | 1.2.3.8 | Pyridoxal to Pyridoxoic acid |
| 9 | Sulfite Oxidase | 1.8.3.1 | Converts Sulfite to Sulfate S2O32- to SO42-) |
| 10 | Biotin Sulfoxide Reductase | - | Converts Biotin Sulfoxide to Biotin |
| 11 | Dimethyl Sulfoxide Redutase | 1.8.5.3 | Converts Dimethyl Sulfoxide to Dimethyl sulfide [(CH3)S=O to (CH3)2S] |
| 12 | Tetrathionite Reductase | - | Converts Tetrathionite to Thiosulfite (S4O62- to S2O32-) |
Table 1: Enzyme commission numbers and functions of all molybdenum containing enzymes mentioned in this mini-review.
Nitrogenase, isolated from N2-fixing bacteria, is the enzyme catalyzing the reduction of molecular N2 to ammonia (NH3). In addition to the reduction of molecular nitrogen N2, Nitrogenase can also reduce a variety of small, unsaturated substrates including the reduction of C2H2 to C2H4, N2O to N2 and H2O, N3- to NH4+ and N2, and HCN to CH3NH2 [17,18]. Nitrogenase is composed of two proteins, dinitrogenase (FeMo protein) and dinitrogenase reductase (Fe protein). Dinitrogenase reductase is a dimer of two identical α2 subunits that bridge one ferredoxin-like (Fe4S4)2+/1+ cluster. Dinitrogenase is a α2β2 tetramer, and it carries a unique iron-molybdenum cluster that contains Fe, Mo and S with one cagelike MoFe7S9 homocitrate cluster per α, β-pair. It is believed that the substrate molecules (such as molecular nitrogen, N2) bind inside this FeMo cluster [4-8]. The Mo center is octahedrally coordinated to three sulfides (μ3-S ligands), a histidine imidazole side chain, and two oxygen atoms from homocitrate. Supplementary Figure 2 shows the structure formula of the active site of the FeMo cofactor in the N2 fixing enzyme; Nitrogenase.
Burris and coworkers demonstrated that citrate substitutes for homocitrate in nitrogenase of a nifV mutant of Klebsiella pneumonia [9]. In dinitrogenase, the homocitrate ligand is coordinated to the Mo center via the central carboxyl and the central hydroxyl group. We have visited this subject a decade ago with our detailed speciation article [19]. Molybdenum is believed to be taken up by organisms as the molybdate anion MoO42-. This would be essential for the biosynthesis of the final FeMo cofactor from an oxomolybdenum citrate precursor [13-16]. Tricarboxylic acids, citric and homocitric acids may play an essential role in the mobilization of molybdenum during cofactor biosynthesis and the mobilized oxomolybdenum tricarboxylate fragment must undergo reduction, exchange the oxo ligands for the sulfide ligands, and merge with the rest of the nifB. While the precise role of tricarboxylic acids in the biosynthesis of the FeMo cofactor and the mechanism of dinitrogenase reduction is still regarded as poorly understood [15], the elucidation of the role played by the tricarboxylic acids has been pursued with great interest [9-16,19].
Many researchers, including ourselves, have attempted to mimic the reaction of Mo6+ with that of mono-hydroxyl poly-carboxylates (citrate, homocitrate and malate) [9- 16,19-23]. Herein, we show the outcome of the reactions of the simple mono-hydroxyl di-carboxylate (Malate) in the form of the protonated Malic acid (H2MA) with Mo6+ in aqueous solutions under ambient conditions in 0.1 M ionic strength (NaNO3).
Experimental Section
Materials and method
Aqueous solutions of D,L-Malic acid (H2MA) were prepared using 99% purity (Sigma reagent grade), formula weight 134.09 g.mol-1. Aqueous molybdenum (Mo6+) solutions were prepared from the crystalline ammonium salt of molybdic acid, (NH4)6Mo7O24.4H2O, 83% MoO3, formula weight 1235.9 g.mol-1, also Sigma reagent grade. The primary (1°) standard potassium hydrogen phthalate (KHP, 99.99%) and the solid sodium hydroxide pellets (NaOH, 98%) were purchased from Fisher Chemical Company. We have standardized the sodium hydroxide (NaOH) solution to the fourth digits to the right of the decimal point. All pH values were measured using Thermo-Orion Membrane Advanced ISE/pH/mV/ORP meter (model 720A+) connected to the accurate-combination Orion-glass electrode in 0.1 mol.L-1 ionic strength. The 0.10 M ionic strengths of all solutions were adjusted by the addition of 10% v/v of 1.0 M NaNO3 solution.
Potentiometry
The detailed methods used to carry potentiometric titrations are well established in the literature [19,21- 24]. For each individual experiment, H2MA solutions were first added to the titration vessel, followed by the addition of the specified amount of Mo6+ solution. The final total volume of the reacting species was 100 mL. NaOH solution was added in 100 μL increments by means of the calibrated and accurate, 100 μL total capacities, Eppendorf micro-pipette. Each individual potentiometric titration took about 3.5 h to complete.
Results and Discussion
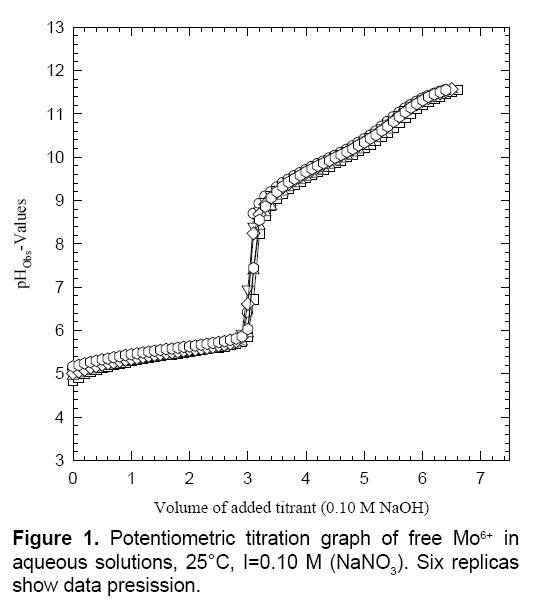
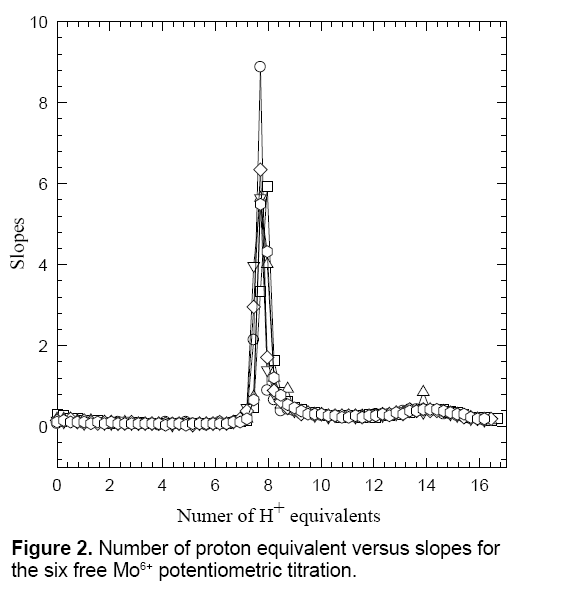
Figure 1 shows the potentiometric titration plots of free Mo6+. The free H2MA titration curve has been published in previous studies [24,25-27]. It is established that H2MA releases two protons out of the two carboxylic acid groups. Additionally, the hydroxyl group can participate in metal ion chelation to form two fused six-membered and five-membered very robust ring chelates [19,25-27]. Figure 2 shows the potentiometric titration plots of Mo6+ in the form of number of proton equivalents released (x-axis) versus the first derivatives or slopes (y-axis) of the experimental pH-values (the inflection points appeared at 7.78 ± 0.10, n=6). This observation parallels that of the previous study of Mo6+ with citric acid which is consistent with the literature values [19].
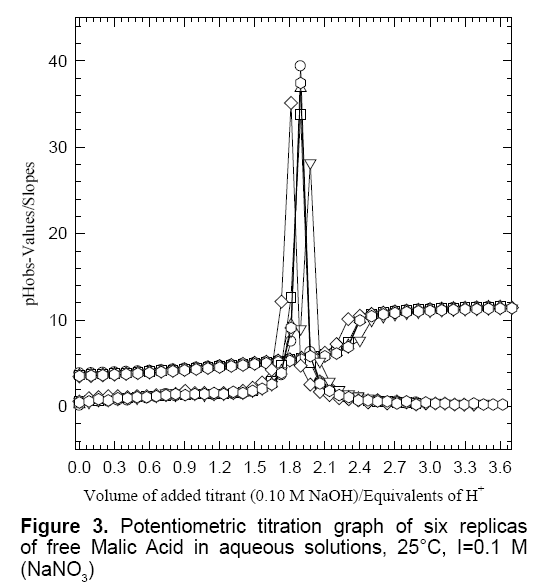
Figure 3 is the double plots of free H2MA in which the potentiometric titrations of six replicas in the form of volume of added titrant versus experimental pH-values are overlaid with the number of proton equivalents on the x-axis versus the slopes on the y-axis. A similar titration plateau was observed in our previous studies [25,27]. This particular titration graph is the very same graph that has been collected alongside the reaction of the hexavalent molybdenum (Mo6+) in its reaction with malic acid (H2MA); see below for details of the Mo6+ reaction with H2MA.
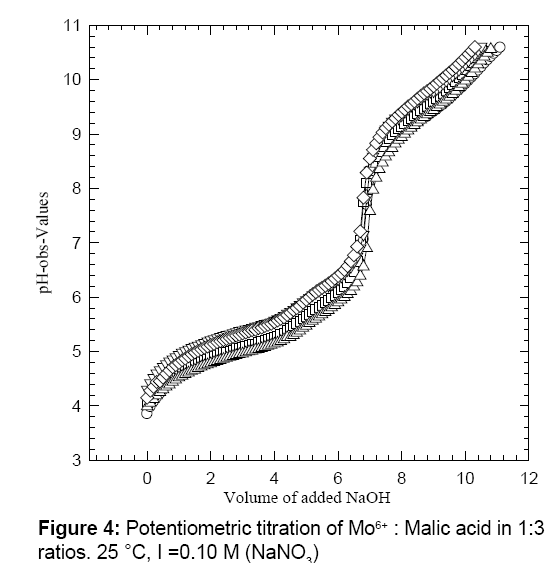
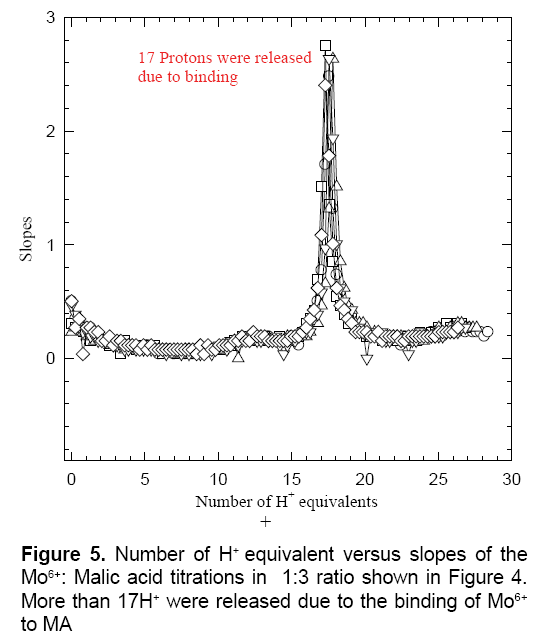
Figure 4 shows the titration plateaus of the reaction mixture of Mo6+ with MA in 1:3 M ratios. Excess amount of malic acid was selected for three reasons: (1) to avoid metal ion hydrolysis; (2) to avoid the formation of any oligomeric metal-complexes that were formed when the ratio of the Mo6+ ion was in excess [21-23]; and (3) for ease of pipetting 2.00 mL H2MA with 2.00 mL Mo6+, which gave this 1:3 molar reaction ratios. After calculating the number of proton equivalents of the reactions of Mo6+ with H2MA in 1:3 M ratios (Figure 5), it appeared that ~ 17 proton equivalents were released into the solution (17.01 ± 1.22, n=6). This observation is in a very good agreement that one might expect for the release of a net 7.78 proton equivalents from the free mole of Mo6+ in addition to the net of nine protons total out of the three available protons to be released per malic acid molecule (i.e., 9H+ from the three moles of H2MA plus 7.78 from the free Mo6+ ion). In n Figure 5, it shows the values which are equal to 16.78 proton equivalents.
Figure 5 confirms that the malic acid is releasing three H+ equivalents although it is referred to as H2MA in the literature [19,24-27]. This is due to the central hydroxyl group upon chelation to Mo6+ (or any metal ion for that matter) releases its proton. It is concluded from this data that the main complexes that have been formed in solution are those according to equation 1.
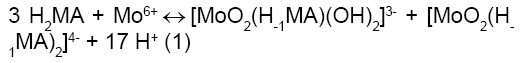
Conclusion
The current experimental data presented in this mini-review is novel. When the detailed literature reviews were searched for the reactions of H2MA with the hexavalent molybdenum ion Mo6+, only few papers were found [21-23]. In these studies, they all agreed that the aqueous solution chemistry of Mo6+ is not that simple. They also agreed on the formation of the simple one to one complexes [MoO2(MA) (OH)2]3-, [MoO2(MA)2]4-, [MO2O5(MA)2(OH)2]4- with stability constant values of Logβ of 8.17, 13.89 and 22.31, respectively [22]. When the crystal structure was published two years later after these initial aqueous attempts in 1983 [21], it turned out that what crystallized was the bis-mono-protonated complex which has the following molybdenum malate complex formula: [MoO2(Hmal)2]2- [21]. We do not believe that we are observing this bis-mono-protonated complex. What concerns us here is not whether we are observing the formation of the di- anion, the tri-anion, or the tetra-anion molybdenum-malate complex. We are also not concerned with whether it is the monoprotonated or the de-protonated or the (hydroxo) molybdenum-malate complex. However, in this study the significant revelation is that we have identified the total number of proton equivalents released into the solution which gives the same binding mode as seen in the crystal structure and the previous equilibrium studies [21-24].
The detailed and the most trusted reviews of the molybdoenzymes by researchers [1-3] stressed that the bi-dentate binding mode of the monohydroxyl poly-carboxylates, whether it is in the form of citrate or homocitrate, is the dominant mode of binding. Here in, we are stressing that the formed complex of Mo6+ with malic acid released a net of 17 proton equivalents which can only be accounted for by the binding of Malate in a bi-dentate or a tridentate fashion (the potentiometric titration is lacking supplying this information). The stoichiometry given in the equilibrium shown in equation (1) is only accounted for by the formation of a mixture of the two proposed molybdenum-malate complexes depicted, i.e., [MoO2(H-1MA)(OH)2]3- + [MoO2(H-1MA)2]4-. This mixture of the two complexes released a net of 17 H+. It is noteworthy that these two complexes are consistent with the ones identified by others [21-24].
Acknowledgement
We would like to acknowledge the financial support from NSF under Grant # HRD-1332459. Special thanks go to Professor Sherry Painter for reading the manuscript and her many helpful suggestions. of awards and prizes.
References
- Burgmayer SJN, Stiefel EI. (1985). Molybdenum Enzymes, cofactors and model systems. J Chem Educ. 62: 943-953.
- Hille R, Hall J, Basu P. (2014). The mononuclear molybdenum enzymes. Chem Rev. 114: 3963-4038.
- Swedo KG, Enmark JJ. (1979). Some aspects of the bioinorganic chemistry of molybdenum. J Chem Educ. 56: 70-76.
- Kim J, Rees DC. (1992). Structural models for the metal centers in the nitrogenase molybdenum-iron protein. Science. 257: 1677-1682.
- Kim J, Rees DC. (1992). Crystallographic structure and functional implication of the nitrogenase molybdenum-iron protein from A. vinelandii. Nature. 360: 553-560.
- Kim JM, Woo D, Rees DC. (1993). X-ray crystal structure of the nitrogenase molybdenum-iron protein from Clostridium pasteurianum at 3.0 Å resolution. Biochemistry. 32: 7104-7115.
- Chan MK, Kim J, Rees DC. (1993). The nitrogenase Fe-Mo-cofactor and P-Cluster pair. 2.2 Å resolution structures. Science. 260: 792-794.
- Kim J, Rees DC. (1994). Nitrogenase and biological nitrogen fixation. Biochemistry. 33: 389-398.
- Burris RH, Liang J, Madden M, et al. (1990). Citrate substitutes for homocitrate in nitrogenase of a nifV mutant of Klebsiella pneumonia. Biochemistry. 29: 8577-8581.
- Zhou ZH, Wan HL, Tsai KR. (2000). Synthesis and spectroscopic and structural characterization of molybdenum(VI) citrato monomeric raceme and dimer, K4[MoO3(cit)] 2H2O, the dinuclear K4[(MoO2)2O(Hcit)2] 4H2O. Inorg Chem. 39: 59-64.
- Shah VK, Allen JR, Spangler NJ, et al. (1994). In vitro synthesis of the iron-molybdenum cofactor of nitrogenase. Purification and characterization of NifB cofactor, the product of NIFB protein. J Biol Chem. 269: 1154-1158.
- Wright DW, Chang RT, Mandal SK, et al. (1996). A novel vanadium(V) homocitrate complex: Synthesis, structure and biological relevance of [K2(H2O)5][(VO2)2(R,S-homocitrate)2]. J Bioinorg Chem. 1: 143-151.
- Gronberg KLC, Gormal CA, Durrant MC, et al. (1998). Why R-Homocitrate is essential to the reactivity of FeMo-cofactor of nitrogenase: Studies on NifV-extended FeMo-cofactor. J Am Chem Soc. 120: 10613-10621.
- Pedrosa de Jesus JD, Farropas MdeD, O'Brien P, et al. (1983). Photochemical studies of Mo(VI) citric acid complex MO2O5OH(H2O)(C6H5O7)2-. Trans Me Chem. 8: 193-195.
- Cruywagen JJ, Van de Water RF. (1986). Complexation between molybdenum and citrate: A potentiometric and calorimetric investigation. Polyhedron. 5: 521-526.
- Cruywagen, JJ, Rohwer EA, Wessels GFS. (1995). Molybdenum(VI) complex formation-8. Equilibria and thermodynamic quantities for the reaction with citrate. Polyhedron. 14: 3481-3493.
- Lippard, SJ, Berg JM. (1994). In Principles of Bioinorganic Chemistry, University Science Books, Mill Valley, CA.
- Cowan JA. (1997). In Inorganic Biochemistry/An introduction, Wiley-VCH Inc. New York, NY.
- Hamada YZ, Bayakly N, George D, et al. (2008). Speciation of Mo(VI)-Citrate complexes in aqueous solutions. Synthesis and Reactivity of Inorganic and Metal-Organic and Nano-Metal Chemistry. 38: 664-668.
- Zhou ZH, Yan WB, Wan HL, et al. (2002). Synthesis and characterization of homochiral polymeric S-malato molybdate(VI): Toward the potentially stereospecific formation and absolute configuration of iron-molybdenum cofactor in nitrogenase. J Inorg Biochem. 90: 137-143.
- Carolyn BK, Arran JW, Richard NH, et al. (1983). Molybdenum (VI) complexes with malic acid: Their inter-relationships and the crystal structure of dicaesium bis[(S)-malato(2-)]-cis-dioxomolybdate(VI)-Water(1/1). J Chem Soc. 7: 1299-1303.
- Aurelio B, Antonio CA, José B. (1981). Study of the complexes of Mo(VI) with malic acid. J Inorg Nucl Chem. 43: 1337-1341.
- Miloš B, Josef H, Dalibor M. (1986). Chelates of molybdenum(VI) with citrate and malate. Collect Czech Chem Commun. 51: 2702-2711.
- Martell AE, Smith RM, Motekaitis RJ. (2001). Critical stability constants database, Version 6.0, NIST, Texas A & M University, College Station, TX, USA.
- Hamada YZ, Harris M, Burt K, et al. (2013). Detailed potentiometric study of Al3+ and Cr3+ with malic acid in aqueous solutions. Journal of Membrane and Separation Technology. 2: 1-6.
- Hamada YZ, Bayakly N, Peipho A, et al. (2006). Accurate potentiometric studies of chromium-citrate and ferric-citrate complexes in aqueous solutions at physiological and alkaline pH values. Syn React Inorg Metal Org Nanometal Chem. 36: 469-476.
- Hamada, YZ, Carlson B, Dangberg J. (2005). Interaction of lactate and malate with chromium(III) and iron (III) in aqueous solutions. Syn React Inorg Metal Org Nanometal Chem. 35: 515-522.

Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences